Open data framework for biology
Query, trace, and validate datasets and models at scale. Automate context for agents and humans.
One API: lakehouse, lineage, feature store, ontologies, bio-registries & formats.
From the creators of

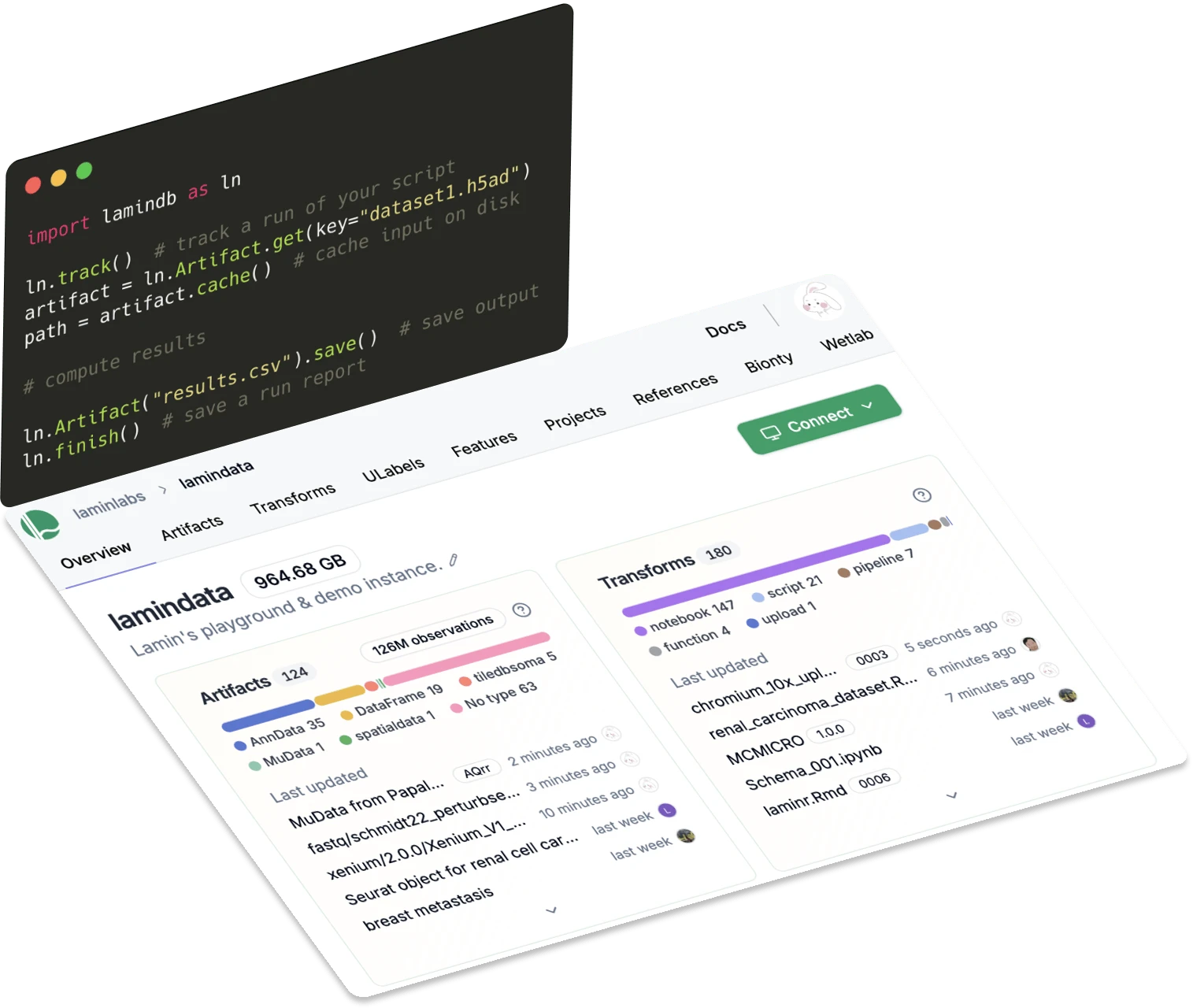
Lineage
Trace data, code & reports
Know where data came from and what it's used for. Track data lineage with a single line of code.
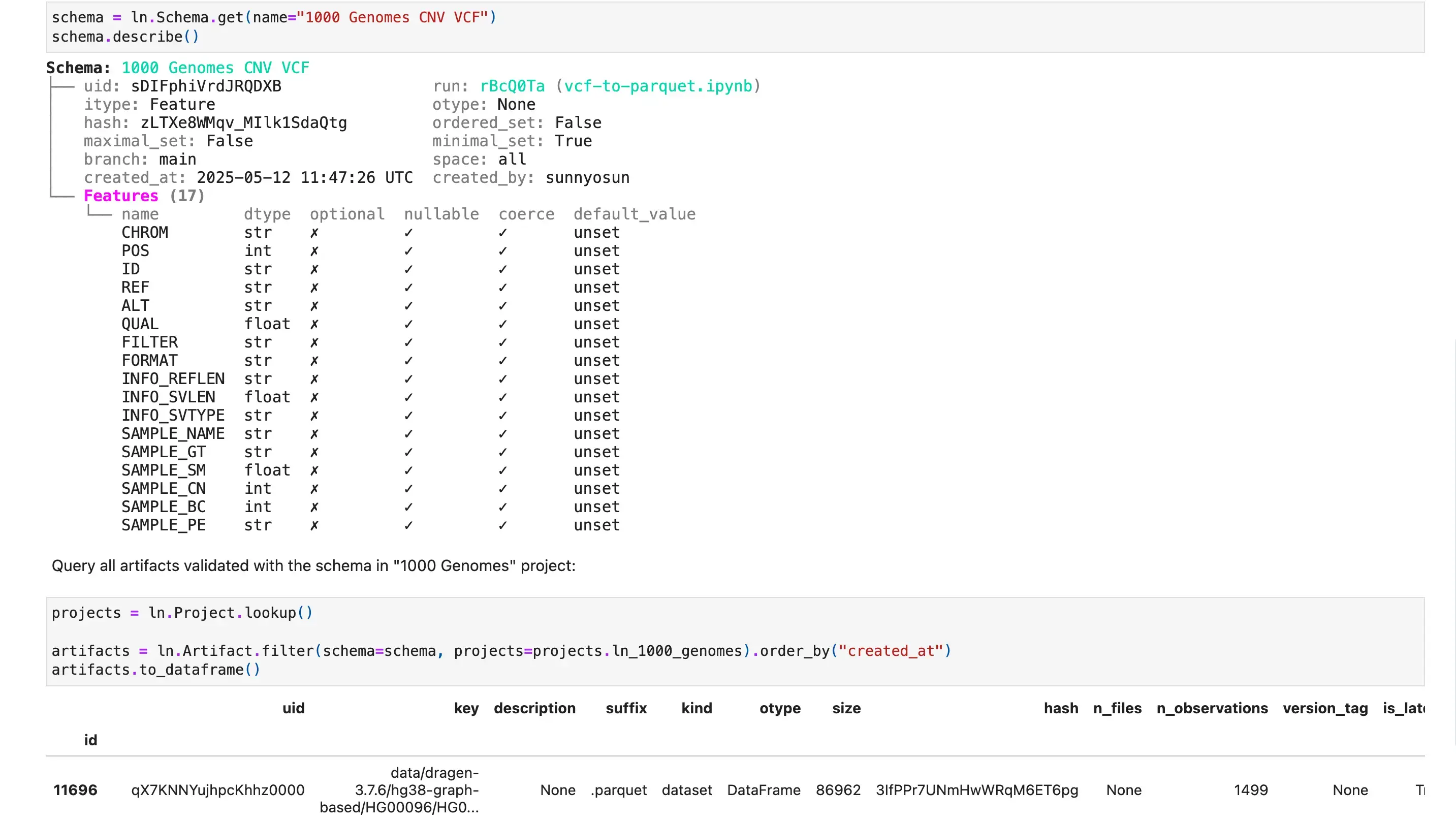
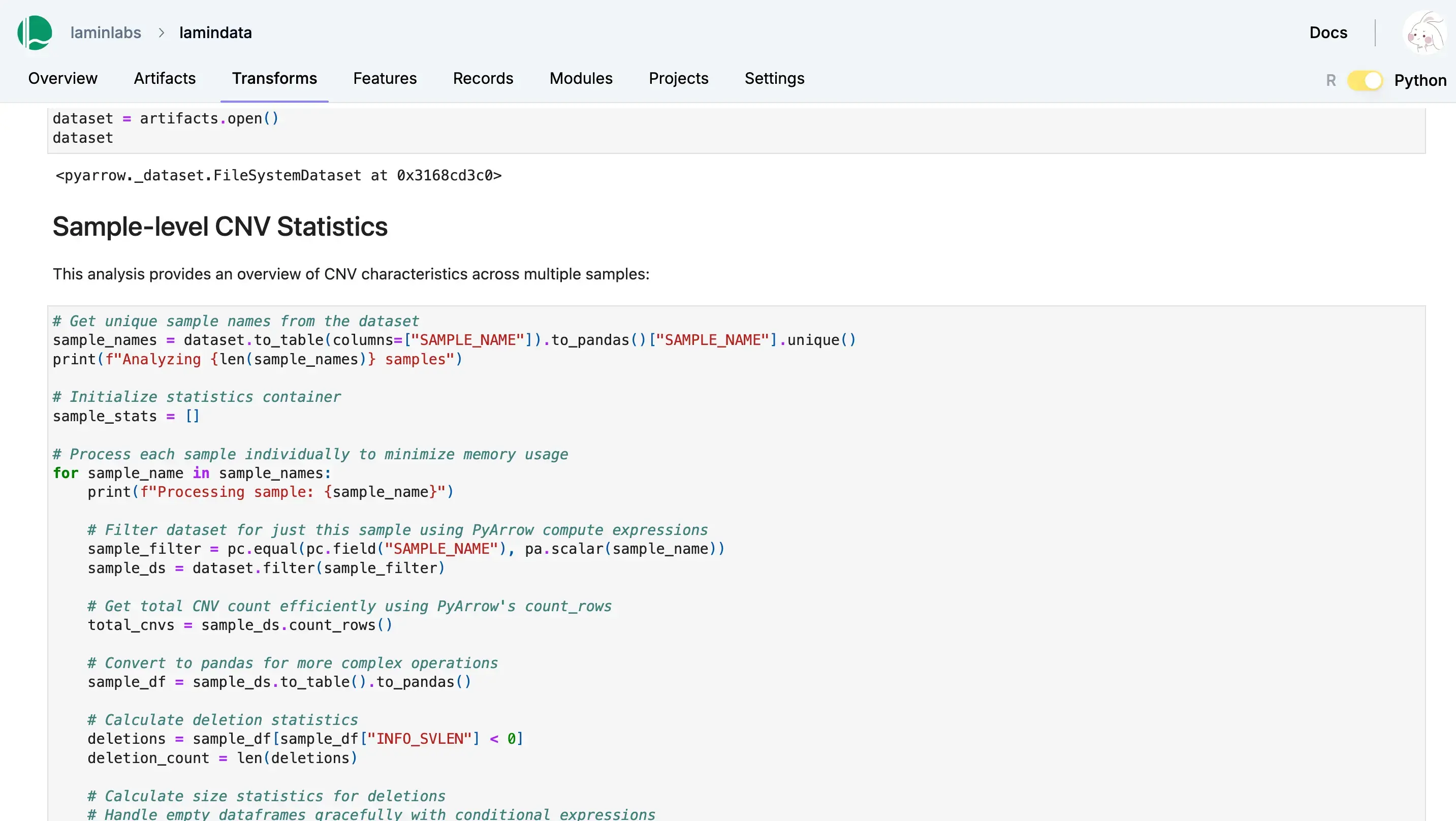
Lakehouse + Bio-formats
Query datasets at scale
Query and batch-load datasets with lakehouse support for a wide range of table & array formats. Manage their features & schemas as metadata in Postgres or SQLite.
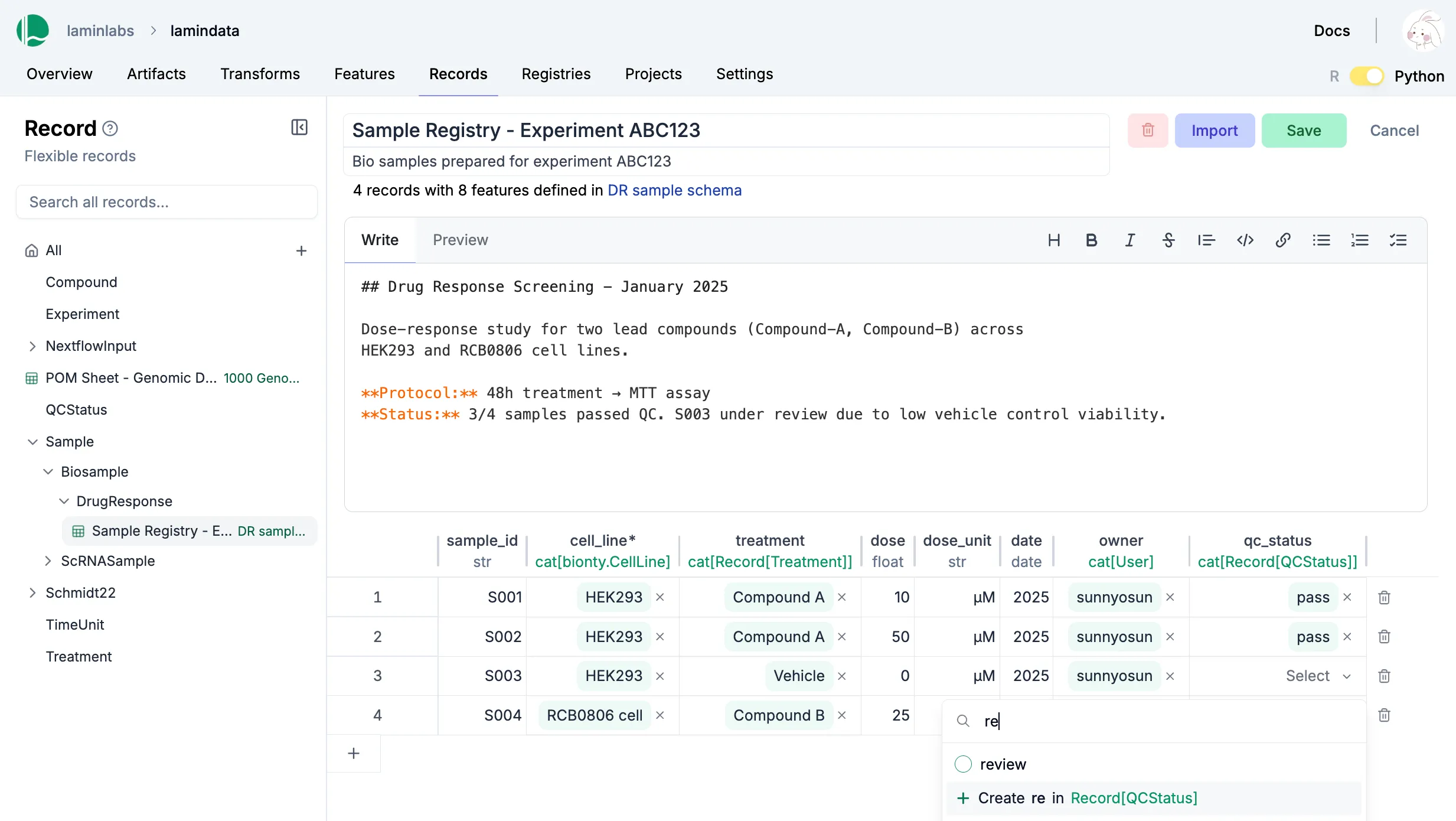
Registries + Sheets (LIMS)
Unify metadata and datasets
Manage metadata in relational sheets that maps directly to data in storage. A single Python/R class with built-in ontologies, project & change management.
Integrity
Validate & annotate datasets
Use schemas to enforce consistency across your data assets. Annotate datasets with a single line of code.
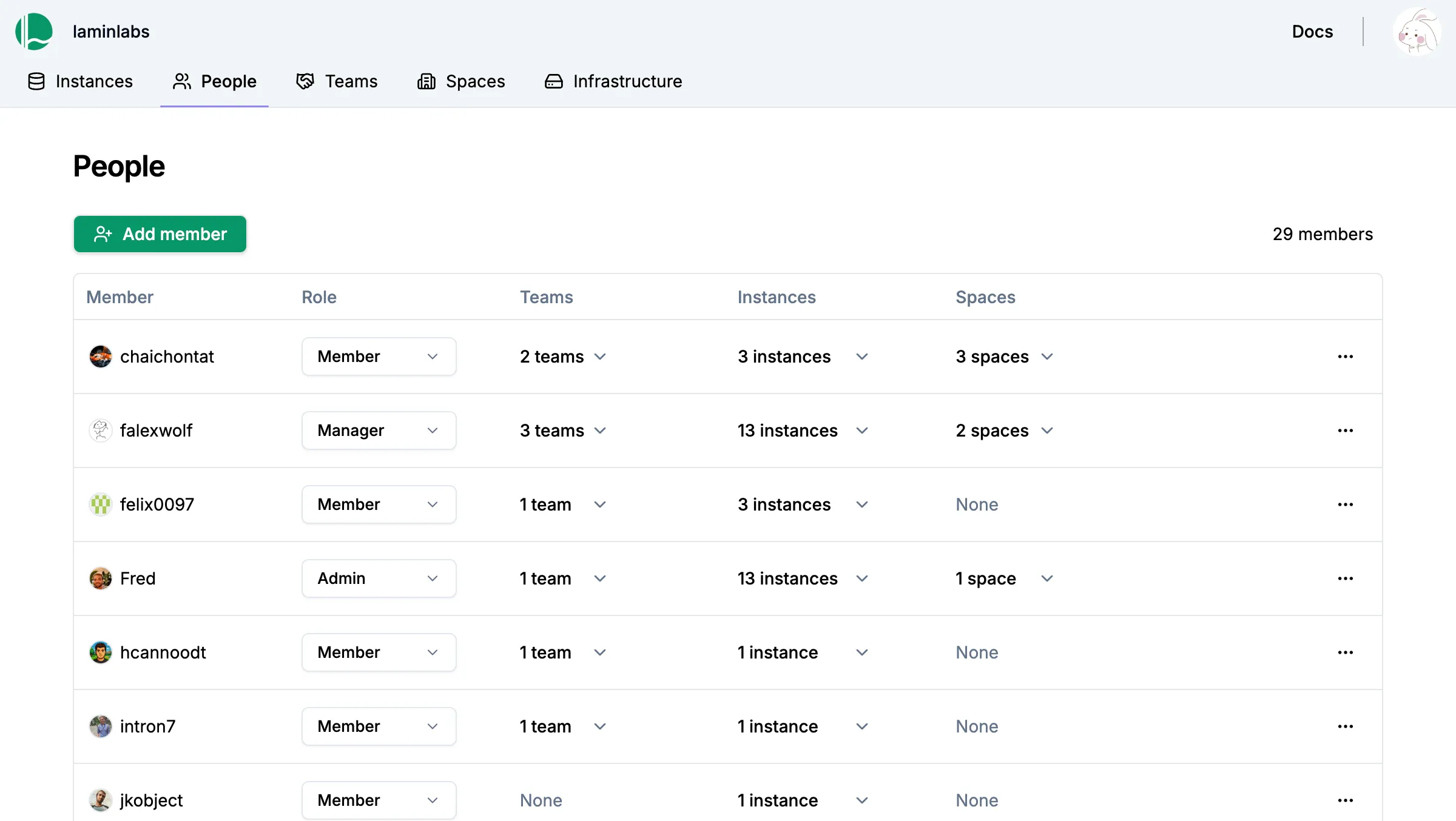
Zero lock-in
Administrate with ease while staying in control
Manage fine-grained access for humans & agents with SaaS-like simplicity. Enjoy full admin control at the database and storage level.
Context
Build your organization's long-term memory
As your team and agents work, data, models & reports are automatically linked and mapped into queryable feature spaces — building context & training data that compounds over time.