- Docs · Repository · Tweet · LinkedIn · DOI
- ⸻ 2022-08-29
nbproject: Manage Jupyter notebooks#
Abstract#
nbproject is an open-source Python tool to help manage Jupyter notebooks with metadata, dependency, and integrity tracking. A draft-to-publish workflow creates more reproducible notebooks with context.
There are a number of approaches to address reproducibility & manageability problems of computational R&D projects. nbproject complements - and should be combined with - approaches that are based on modularizing notebooks into pipelines, containerizing compute environments, or managing notebooks on centralized platforms.
Introduction#
Over the past 11 years, Jupyter notebooks [Pérez07, Kluyver16] have become data scientists’ most popular user interface [Perkel18].[1] Today, GitHub hosts about 9M publicly accessible notebooks in active repositories at exponential growth (Figure 1). VS Code made notebooks an integral component of its developer experience [Dias21] and many cloud services offer a workbench built around Jupyter Lab.
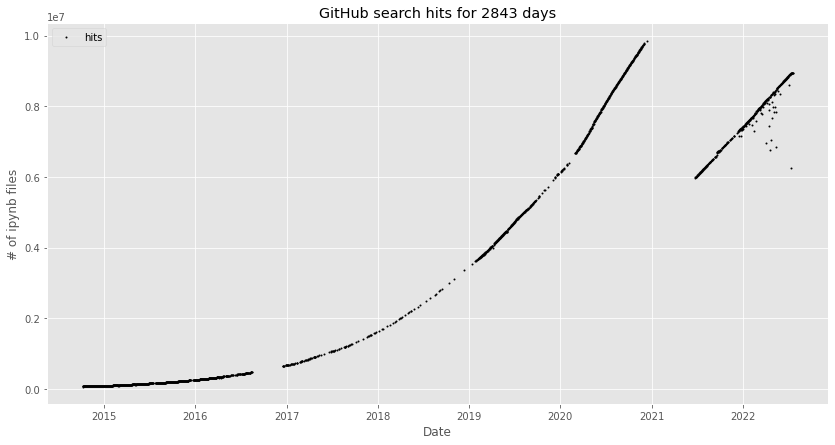
Figure 1: Number of public GitHub-hosted Jupyter notebooks. The graph is from nbestimate and was generated in July 2022 [Parente22] (MIT license). The kink in December 2020 was caused by GitHub changing their query results to exclude repositories without activity for the past year.#
Nonetheless, the average notebook-based data science workflow has a severe reproducibility problem [Perkel18, Balogh22].[2] In addition, large notebook-based projects are hard to manage and often develop into an organic collection of notebook files that are hard to navigate. The situation seems particularly severe in biology, where even scientific results that are published in high impact journals often come with disorganized, hard-to-reproduce collections of notebooks.
Problems of notebooks#
The overarching project is an unstructured collection of notebooks, code, and data files.
Data dependencies are missing.
Pipeline dependencies (previous data transformations) are missing.
Notebook has low code quality [Grotov22].[5]
Why notebooks, then?#
Notebooks are highly popular across the full breadth of computational sciences & engineering.[6] Notebooks are the standard for prototyping and analysis, while ML Ops tools (see Appendix) are the standard for optimizing narrow classes of models.
Optimizing narrow classes of models though typically doesn’t yield the biggest gains towards progress in computational biology. Computational biology often stops optimizing ML models at stages that other fields would consider prototyping[7] and has always been data-centric [Ng22] through its tight dependence of computational (drylab) on wetlab experiments. Between wetlab experiments, data generation conditions often change so drastically that data scientists’ greatest efforts are spent on assembling & cleaning the data. Hence, instead of optimizing a narrow class of computational models, computational biologists need to continuously conceive new ways of taming and modeling data.
Notebooks’ “computational narrative” offers a format for this type of work: A “document that allows researchers to supplement their code and data with analysis, hypotheses and conjecture”, according to Brian Granger [Peres18]. “Notebooks are a form of interactive computing, an environment in which users execute code, see what happens, modify and repeat in a kind of iterative conversation between researcher and data. […] Notebooks allow more powerful connections between topics, theories, data and results”, according to Lorena Barba [Peres18].
Existing solutions to problems#
Today, problems 5 & 6 are addressed to varying degrees by notebook platforms that allow using notebooks in ML pipelines and help with decomposing them into smaller code modules. Examples for this are Elyra [Resende18] (Figure 2), Ploomber [Blancas20, Blancas21a, Blancas21b] and Orchest [Lamers21]. The latter two and most other notebook platforms (Appendix) also manage (package) computational environments to execute notebooks, and with that, address problem 3.
An interesting alternative approach to making notebooks more reproducible consists in storing the history of users’ actions, as offered by Verdant [Kery19a, Kery19b].
To the authors’ knowledge, all other notebook platforms (Appendix) do not focus on reproducibility and manageability, but on the allocation of compute & storage resources.
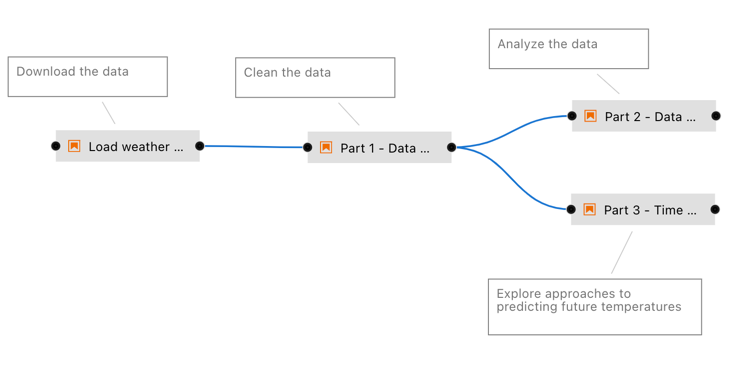
Figure 2: Example for an Elyra notebook pipeline. From the Elyra examples repository, MIT licensed [Resende18].#
Solutions chosen by nbproject#
nbproject complements pipeline and compute environment packaging solutions: nbproject is a lightweight Python package that works without connecting to any centralized platform. It addresses problems as follows:
Project is an unstructured collection of notebooks, code, and data files. nbproject allows configuring arbitrary project- and management-related metadata. Its id and version fields allow anchoring notebooks in a graph of R&D operations of an entire team.
Notebook cells aren’t consecutively executed. nbproject provides a publishing workflow that checks for consecutiveness & the presence of a title and versions the notebook.
Package dependencies are missing. nbproject offers a visual way to track packages, which complements packaging solutions.
Data dependencies are missing. nbproject integrates well with LaminDB.
Pipeline dependencies are missing. nbproject allows sketching pipelines manually. When integrated with LaminDB, it provides full provenance automatically.
Notebook has low code quality. The publishing workflow encourages small modular notebooks with most code residing in versioned packages.
Design choices#
nbproject’s two most important features are tracking & displaying metadata and offering a publishing workflow.
Tracking & displaying metadata#
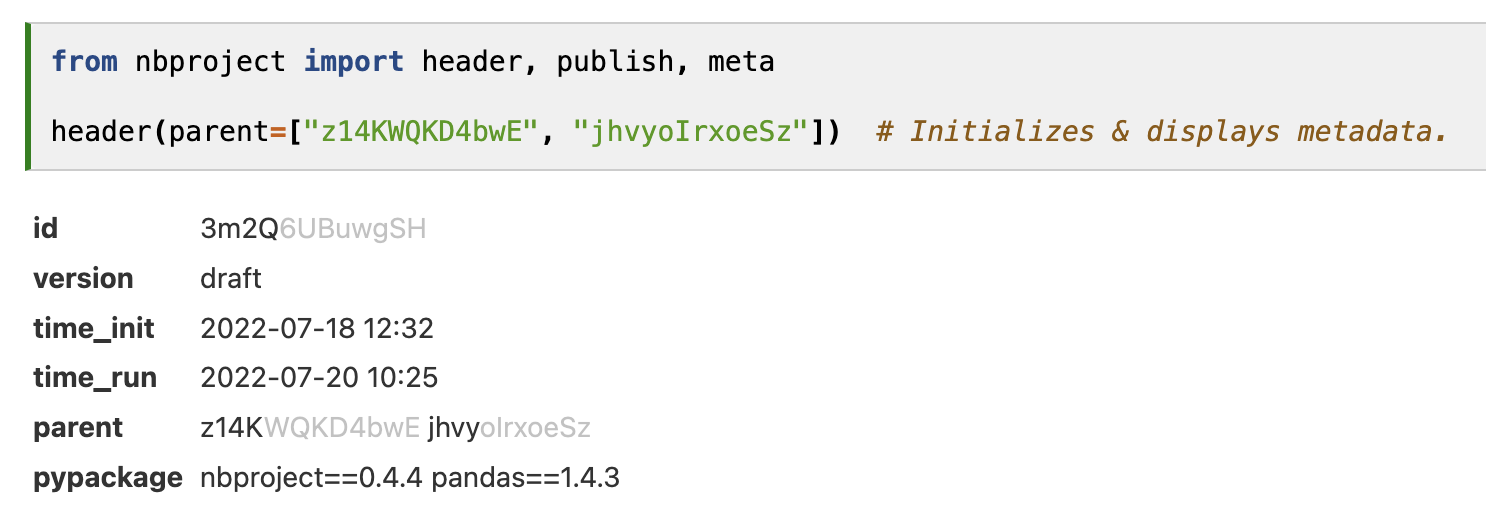
The header (Figure 3) is inspired by popular notebook tools, like Notion, ELNs in biology, and Jupyter notebook platforms. Any user opening their own or someone else’s notebook is provided with relevant context for interpreting the notebook content: a universal ID, a version, time stamps, important dependencies, and arbitrary additional metadata. While most notebook platforms also provide IDs and other metadata for notebooks (Appendix), to our knowledge, only nbproject offers an API to access such metadata.
The metadata header display depends on the computing environment in which the notebook is run: If the environment differs from the stored package dependencies, mismatching versions can readily be seen as both stored and live dependencies are displayed. This typically occurs when multiple users collaborate on notebooks and the receiver of a notebook tries to re-run the notebook of a sender (Figure 3).
Sender |
Receiver |
|---|---|
|
|
The publishing workflow#
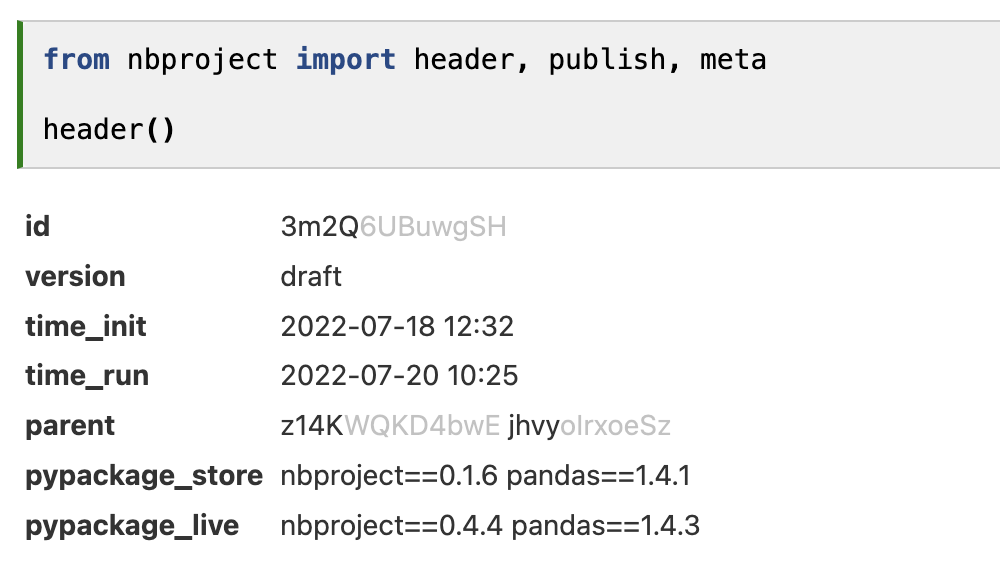
nbproject suggests publishing notebooks before sharing them with a receiver (Figure 4). After an editing phase in “draft” mode, publishing the notebook writes dependencies to the metadata store, checks for the integrity of the notebook (consecutiveness & title), and sets a version number.
Hence, if a user receives a published notebook, they know that the latest dependencies are stored within it, are provided with a version number, and see whether the notebook was consecutively executed.
Figure 4: Publishing workflow. Made with Kat by Sunny Sun.#
Semantic vs. full dependency tracking#
The visual display of mismatching package dependencies between compute environments should be seen in the broader context of reproducibility vs. determinism. Reproducibility is “the ability of an independent research team to produce the same results using the same method based on the documentation made by the original team” (adapted from [Gunderson18]). Computational determinism, in addition, requires the bit-exact same output for the same input. Whereas targeted experimentation or the certification of models for sensitive application areas should run in deterministic environments [Heumos22], reproducibility is sufficient for many applications.
Highly deterministic environments can be created with Docker & conda and managed on most data platforms. However, they don’t ensure scientific correctness of results. In fact, deterministic environments can lead to results that are only reproducible on highly specific environments, say, if they depend on certain data type precisions or greedy algorithms. If a team always uses the exact same environments they will never learn that such a result is indeed only bit-wise correct, but not scientifically correct. The scientific correctness of a robust statistical result, by contrast, should reproduce in many different compute environments as long as basic conditions are met.[8]
Hence, policies in R&D teams may vary from using the same Docker container for all computations to making Docker containers for each analysis to not using Docker at all and running analysis on a variety of environments, cross-checking them among team members for robustness.
Testing notebooks in CI#
Just like software, notebooks should be tested in continuous integration (CI).
As nbproject needs to communicate with a Python kernel, a server, and the frontend of Jupyter editors, we couldn’t use nbmake and related existing testing infrastructure:
nbproject-test.
Instead of relying on the availability of a server in CI environments, nbproject-test executes notebooks with nbclient, infers notebook paths and passes these as environment variables to nbproject.
Acknowledgements#
We are grateful to Sunny Sun & Alex Trapp for providing early feedback and to Sunny for making the GIF of Figure 4.
We are grateful to the Jupyter project developers, in particular, Jeremy Tuloup for ipylab.
Citation#
Cite the software and this report as:
Rybakov S, Heumos L & Wolf A (2022). nbproject: Manage Jupyter notebooks. Lamin Blog. https://doi.org/10.56528/nbp
Appendix#
General ML Ops tools#
This is a non-comprehensive list of ML Ops tools centered around helping to organize the model development & deployment cycle.
Most of these track data storage, a model registry, and offer ways of creating pipelines.
MetaFlow: A framework for real-life data science.
dvc: Open-source Version Control System for Machine Learning Projects with strong git inspiration.
Neptune AI: Experiment tracking and model registry for production teams.
Kedro: Kedro is an open-source Python framework for creating reproducible, maintainable and modular data science code.
MLFlow: An open source platform for the machine learning lifecycle.
Zen ML: Build portable, production-ready MLOps pipelines.
dstack: Your new home for building AI apps.
Dedicated notebook platforms#
Solutions that offer building pipelines or experiment tracking based on notebooks, as discussed in the main text:
Ploomber: Data & ML Ops platform. For notebooks, it uses ipytext to turn notebooks into diffable code files, and allows creating pipelines.
Orchest: Data & ML Ops platform. Offers pipelines for notebooks and scripts.
Verdant: Track user edit histories of notebooks.
Elyra: Pipelines for notebooks and scripts.
Solutions offering compute allocation, storage connection, and environment management:
Notebook tracking in data platforms, with provenance features typically through pipelines are provided by leading data platforms: Databricks (MLFlow), Snowflake, Domino Data Labs, Palantir, Cloudera.
Projects related to managing notebooks, some of them mentioned in a popular blog post from Netflix [Ufford18]:
Bookstore: Notebook storage and publishing workflows for the masses. No longer maintained.
Commuter: Notebook sharing hub based on an nteract server.
nbss: Notebook sharing hub.
nbdev: Turn notebook projects into Python development projects.
Papermill: Parametrize & execute Jupyter notebooks at scale.
ML Exchange: Open source data platform with notebook support.
ExecutableBooks: Publish notebooks with metadata.
Metadata tracking#
There are two existing open-source projects concerned with general metadata management for notebooks: nbmetalog [Moreno21] and nbmeta. While the latter seems no longer maintained, the former provides a convenient way to access session & execution metadata about the notebook.
There were suggestions for assigning IDs to notebooks in Project Jupyter itself (see here). IDs for notebook cells are meanwhile a standard.
Google Colab provides an ID, time stamps, provenance & authorship metadata like this:
"metadata": {
"colab": {
"name": "post_run_cell",
"provenance": [
{
"file_id": "1Rgt3Q7hVgp4Dj8Q7ARp7G8lRC-0k8TgF",
"timestamp": 1560453945720
},
{
"file_id": "https://gist.github.com/blois/057009f08ff1b4d6b7142a511a04dad1#file-post_run_cell-ipynb",
"timestamp": 1560453945720
}
],
"provenance": [],
"collapsed_sections": [],
"authorship_tag": "ABX9TyPeUMNWCzl3N44NkFSS3tg0",
"include_colab_link": true
}
...
}
Deepnote provides metadata like this:
"metadata": {
"orig_nbformat": 2,
"deepnote": {
"is_reactive": false
},
"deepnote_notebook_id": "b9694295-92fc-4a3f-98b3-f31609abf37a",
"deepnote_execution_queue": []
}
References#
-
Balogh (2022). Data Science Notebook Life-Hacks I Learned From Ploomber. Machine Learning Mastery Blog.
-
Blancas (2020). Introducing Ploomber. Ploomber Blog.
-
Blancas (2021a). On writing clean Jupyter notebooks. Ploomber Blog.
-
Blancas (2021b). We need a Ruby on Rails for Machine Learning. Ploomber Blog.
-
Cheptsov (2022). Notebooks and MLOps. Choose one. MLOps Fluff.
-
Dias (2021). The Coming of Age of Notebooks. VS Code Blog.
-
Odd Erik Gundersen and Sigbjørn Kjensmo (2018). State of the Art: Reproducibility in Artificial Intelligence. Proceedings of the AAAI Conference on Artificial Intelligence, 32(1).
-
Heumos, Ehmele, Kuhn, Menden, Miller, Lemke, Gabernet & Nahnsen: mlf-core: a framework for deterministic machine learning. arXiv:2104.07651
-
Lamers (2021). Hello, World! Orchest Blog.
-
Resende, Chin, Titzler & Elyra Development Team (2018). Elyra extends JupyterLab with an AI centric approach. GitHub.
-
Grotov, Titov, Sotnikov, Golubev & Bryksin (2022). A Large-Scale Comparison of Python Code in Jupyter Notebooks and Scripts. arXiv:2203.16718.
-
Grus (2018). I don’t like notebooks. YouTube.
-
Jupyter (2018). Jupyter receives the ACM Software System Award. Project Jupyter Blog.
-
Kery (2019). Verdant: A version control tool for JupyterLab that automatically records the history of your experimentation while you work. GitHub.
-
Kery, John, O’Flaherty, Horvath & Myers (2019). Towards effective foraging by data scientists to find past analysis choices. CHI ‘19 #92.
-
Kluyver, Ragan-Kelley, Pérez, Granger, Bussonnier, Frederic, Kelley, Hamrick, Grout, Corlay, Ivanov, Avila, Abdalla, Willing & Jupyter Development Team (2016). Jupyter Notebooks – a publishing format for reproducible computational workflows. Positioning and Power in Academic Publishing: Players, Agents and Agendas 87–90.
-
Perkel (2018). Why Jupyter is data scientists’ computational notebook of choice. Nature 563, 145.
-
Pérez & Granger (2007). IPython: A system for interactive scientific computing. Computing in science & engineering 9, 21.
-
Moreno (2021). nbmetalog helps you log Jupyter notebook metadata. GitHub.
-
Ng & Strickland (2022). Andrew NG: Unbiggen AI. IEEE Spectrum.
-
Parente (2022). Estimate of Public Jupyter Notebooks on GitHub. GitHub.
-
Ufford, Pacer, Seal & Kelley (2018). Beyond Interactive: Notebook Innovation at Netflix. Netflix Tech Blog.