Tutorial: Artifacts#

Biology is measured in samples that generate batched datasets.
LaminDB provides a framework to transform these datasets into more useful representations: validated, queryable collections, machine learning models, and analytical insights.
The tutorial has two parts, each is a Jupyter notebook:
Tutorial: Artifacts - register & access
Tutorial: Features & labels - validate & annotate
Setup#
Install the lamindb Python package:
pip install 'lamindb[jupyter,aws]'
You can now init a LaminDB instance with a directory ./lamin-tutorial for storing data:
!lamin init --storage ./lamin-tutorial # or "s3://my-bucket" or "gs://my-bucket"
💡 connected lamindb: anonymous/lamin-tutorial
import lamindb as ln
ln.settings.verbosity = "hint"
💡 connected lamindb: anonymous/lamin-tutorial
What else can I configure during setup?
Instead of the default SQLite database, use PostgreSQL:
db=postgresql://<user>:<pwd>@<hostname>:<port>/<dbname>
Instead of a default instance name derived from storage, provide a custom name:
name=myinstance
Beyond the core schema, use bionty and other schemas:
schema=bionty,custom1,template1
For more, see Install & setup.
Track a data source#
The code that generates a dataset is a transform (Transform). It could be a script, a notebook, a pipeline or a UI action.
Let’s track the notebook that’s being run:
ln.settings.transform.stem_uid = "NJvdsWWbJlZS"
ln.settings.transform.version = "0"
ln.track()
💡 Assuming editor is Jupyter Lab.
💡 Attaching notebook metadata
💡 notebook imports: lamindb==0.71.0
💡 saved: Transform(uid='NJvdsWWbJlZS6K79', name='Tutorial: Artifacts', key='tutorial', version='0', type='notebook', updated_at=2024-05-01 18:50:22 UTC, created_by_id=1)
💡 saved: Run(uid='gYXIXnIw7o7ys3cRQgEl', transform_id=1, created_by_id=1)
💡 tracked pip freeze > /home/runner/.cache/lamindb/run_env_pip_gYXIXnIw7o7ys3cRQgEl.txt
By calling track(), the notebook is automatically linked as the source of all data that’s about to be saved!
What happened under the hood?
Imported package versions of current notebook were detected
Notebook metadata was detected and stored in a
TransformrecordRun metadata was detected and stored in a
Runrecord
The Transform class registers data transformations: a notebook, a pipeline or a UI operation.
The Run class registers executions of transforms. Several runs can be linked to the same transform if executed with different context (time, user, input data, etc.).
How do I track a pipeline instead of a notebook?
transform = ln.Transform(name="My pipeline", version="1.2.0")
ln.track(transform)
Why should I care about tracking notebooks?
If you can, avoid interactive notebooks: Anything that can be a deterministic pipeline, should be a pipeline.
Just: much insight generated from biological data is driven by computational biologists interacting with it.
A notebook that’s run a single time on specific data is not a pipeline: it’s a (versioned) document that produced insight or some other form of data representation (with parallels to an ELN in the wetlab).
Because humans are in the loop, most mistakes happen when using notebooks: track() helps avoiding some.
(An early blog post on this is here.)
Manage artifacts#
We’ll work with a toy collection of image files and transform it into higher-level features for downstream analysis.
(For other data types: see Data types.)
Consider 3 directories storing images & metadata of Iris flowers, generated in 3 subsequent studies:
ln.UPath("s3://lamindb-dev-datasets/iris_studies").view_tree()
Show code cell output
iris_studies (3 sub-directories & 151 files with suffixes '.csv', '.jpg'):
├── study0_raw_images
│ ├── iris-0337d20a3b7273aa0ddaa7d6afb57a37a759b060e4401871db3cefaa6adc068d.jpg
│ ├── iris-0797945218a97d6e5251b4758a2ba1b418cbd52ce4ef46a3239e4b939bd9807b.jpg
│ ├── iris-0f133861ea3fe1b68f9f1b59ebd9116ff963ee7104a0c4200218a33903f82444.jpg
│ ├── iris-0fec175448a23db03c1987527f7e9bb74c18cffa76ef003f962c62603b1cbb87.jpg
│ ├── iris-125b6645e086cd60131764a6bed12650e0f7f2091c8bbb72555c103196c01881.jpg
│ ├── iris-13dfaff08727abea3da8cfd8d097fe1404e76417fefe27ff71900a89954e145a.jpg
│ ├── iris-1566f7f5421eaf423a82b3c1cd1328f2a685c5ef87d8d8e710f098635d86d3d0.jpg
│ ├── iris-1804702f49c2c385f8b30913569aebc6dce3da52ec02c2c638a2b0806f16014e.jpg
│ ├── iris-318d451a8c95551aecfde6b55520f302966db0a26a84770427300780b35aa05a.jpg
│ ├── iris-3dec97fe46d33e194520ca70740e4c2e11b0ffbffbd0aec0d06afdc167ddf775.jpg
│ ├── iris-3eed72bc2511f619190ce79d24a0436fef7fcf424e25523cb849642d14ac7bcf.jpg
│ ├── iris-430fa45aad0edfeb5b7138ff208fdeaa801b9830a9eb68f378242465b727289a.jpg
│ ├── iris-4cc15cd54152928861ecbdc8df34895ed463403efb1571dac78e3223b70ef569.jpg
│ ├── iris-4febb88ef811b5ca6077d17ef8ae5dbc598d3f869c52af7c14891def774d73fa.jpg
│ ├── iris-590e7f5b8f4de94e4b82760919abd9684ec909d9f65691bed8e8f850010ac775.jpg
│ ├── iris-5a313749aa61e9927389affdf88dccdf21d97d8a5f6aa2bd246ca4bc926903ba.jpg
│ ├── iris-5b3106db389d61f4277f43de4953e660ff858d8ab58a048b3d8bf8d10f556389.jpg
│ ├── iris-5f4e8fffde2404cc30be275999fddeec64f8a711ab73f7fa4eb7667c8475c57b.jpg
│ ├── iris-68d83ad09262afb25337ccc1d0f3a6d36f118910f36451ce8a6600c77a8aa5bd.jpg
│ ├── iris-70069edd7ab0b829b84bb6d4465b2ca4038e129bb19d0d3f2ba671adc03398cc.jpg
│ ├── iris-7038aef1137814473a91f19a63ac7a55a709c6497e30efc79ca57cfaa688f705.jpg
│ ├── iris-74d1acf18cfacd0a728c180ec8e1c7b4f43aff72584b05ac6b7c59f5572bd4d4.jpg
│ ├── iris-7c3b5c5518313fc6ff2c27fcbc1527065cbb42004d75d656671601fa485e5838.jpg
│ ├── iris-7cf1ebf02b2cc31539ed09ab89530fec6f31144a0d5248a50e7c14f64d24fe6e.jpg
│ ├── iris-7dcc69fa294fe04767706c6f455ea6b31d33db647b08aab44b3cd9022e2f2249.jpg
│ ├── iris-801b7efb867255e85137bc1e1b06fd6cbab70d20cab5b5046733392ecb5b3150.jpg
│ ├── iris-8305dd2a080e7fe941ea36f3b3ec0aa1a195ad5d957831cf4088edccea9465e2.jpg
│ ├── iris-83f433381b755101b9fc9fbc9743e35fbb8a1a10911c48f53b11e965a1cbf101.jpg
│ ├── iris-874121a450fa8a420bdc79cc7808fd28c5ea98758a4b50337a12a009fa556139.jpg
│ ├── iris-8c216e1acff39be76d6133e1f549d138bf63359fa0da01417e681842210ea262.jpg
│ ├── iris-92c4268516ace906ad1ac44592016e36d47a8c72a51cacca8597ba9e18a8278b.jpg
│ ├── iris-95d7ec04b8158f0873fa4aab7b0a5ec616553f3f9ddd6623c110e3bc8298248f.jpg
│ ├── iris-9ce2d8c4f1eae5911fcbd2883137ba5542c87cc2fe85b0a3fbec2c45293c903e.jpg
│ ├── iris-9ee27633bb041ef1b677e03e7a86df708f63f0595512972403dcf5188a3f48f5.jpg
│ ├── iris-9fb8d691550315506ae08233406e8f1a4afed411ea0b0ac37e4b9cdb9c42e1ec.jpg
│ ├── iris-9ffe51c2abd973d25a299647fa9ccaf6aa9c8eecf37840d7486a061438cf5771.jpg
│ ├── iris-a2be5db78e5b603a5297d9a7eec4e7f14ef2cba0c9d072dc0a59a4db3ab5bb13.jpg
│ ├── iris-ad7da5f15e2848ca269f28cd1dc094f6f685de2275ceaebb8e79d2199b98f584.jpg
│ ├── iris-bc515e63b5a4af49db8c802c58c83db69075debf28c792990d55a10e881944d9.jpg
│ ├── iris-bd8d83096126eaa10c44d48dbad4b36aeb9f605f1a0f6ca929d3d0d492dafeb6.jpg
│ ├── iris-bdae8314e4385d8e2322abd8e63a82758a9063c77514f49fc252e651cbd79f82.jpg
│ ├── iris-c175cd02ac392ecead95d17049f5af1dcbe37851c3e42d73e6bb813d588ea70b.jpg
│ ├── iris-c31e6056c94b5cb618436fbaac9eaff73403fa1b87a72db2c363d172a4db1820.jpg
│ ├── iris-ca40bc5839ee2f9f5dcac621235a1db2f533f40f96a35e1282f907b40afa457d.jpg
│ ├── iris-ddb685c56cfb9c8496bcba0d57710e1526fff7d499536b3942d0ab375fa1c4a6.jpg
│ ├── iris-e437a7c7ad2bbac87fef3666b40c4de1251b9c5f595183eda90a8d9b1ef5b188.jpg
│ ├── iris-e7e0774289e2153cc733ff62768c40f34ac9b7b42e23c1abc2739f275e71a754.jpg
│ ├── iris-e9da6dd69b7b07f80f6a813e2222eae8c8f7c3aeaa6bcc02b25ea7d763bcf022.jpg
│ ├── iris-eb01666d4591b2e03abecef5a7ded79c6d4ecb6d1922382c990ad95210d55795.jpg
│ ├── iris-f6e4890dee087bd52e2c58ea4c6c2652da81809603ea3af561f11f8c2775c5f3.jpg
│ └── meta.csv
├── study1_raw_images
│ ├── iris-0879d3f5b337fe512da1c7bf1d2bfd7616d744d3eef7fa532455a879d5cc4ba0.jpg
│ ├── iris-0b486eebacd93e114a6ec24264e035684cebe7d2074eb71eb1a71dd70bf61e8f.jpg
│ ├── iris-0ff5ba898a0ec179a25ca217af45374fdd06d606bb85fc29294291facad1776a.jpg
│ ├── iris-1175239c07a943d89a6335fb4b99a9fb5aabb2137c4d96102f10b25260ae523f.jpg
│ ├── iris-1289c57b571e8e98e4feb3e18a890130adc145b971b7e208a6ce5bad945b4a5a.jpg
│ ├── iris-12adb3a8516399e27ff1a9d20d28dca4674836ed00c7c0ae268afce2c30c4451.jpg
│ ├── iris-17ac8f7b5734443090f35bdc531bfe05b0235b5d164afb5c95f9d35f13655cf3.jpg
│ ├── iris-2118d3f235a574afd48a1f345bc2937dad6e7660648516c8029f4e76993ea74d.jpg
│ ├── iris-213cd179db580f8e633087dcda0969fd175d18d4f325cb5b4c5f394bbba0c1e0.jpg
│ ├── iris-21a1255e058722de1abe928e5bbe1c77bda31824c406c53f19530a3ca40be218.jpg
│ ├── iris-249370d38cc29bc2a4038e528f9c484c186fe46a126e4b6c76607860679c0453.jpg
│ ├── iris-2ac575a689662b7045c25e2554df5f985a3c6c0fd5236fabef8de9c78815330c.jpg
│ ├── iris-2c5b373c2a5fd214092eb578c75eb5dc84334e5f11a02f4fa23d5d316b18f770.jpg
│ ├── iris-2ecaad6dfe3d9b84a756bc2303a975a732718b954a6f54eae85f681ea3189b13.jpg
│ ├── iris-32827aec52e0f3fa131fa85f2092fc6fa02b1b80642740b59d029cef920c26b3.jpg
│ ├── iris-336fc3472b6465826f7cd87d5cef8f78d43cf2772ebe058ce71e1c5bad74c0e1.jpg
│ ├── iris-432026d8501abcd495bd98937a82213da97fca410af1c46889eabbcf2fd1b589.jpg
│ ├── iris-49a9158e46e788a39eeaefe82b19504d58dde167f540df6bc9492c3916d5f7ca.jpg
│ ├── iris-4b47f927405d90caa15cbf17b0442390fc71a2ca6fb8d07138e8de17d739e9a4.jpg
│ ├── iris-5691cad06fe37f743025c097fa9c4cec85e20ca3b0efff29175e60434e212421.jpg
│ ├── iris-5c38dba6f6c27064eb3920a5758e8f86c26fec662cc1ac4b5208d5f30d1e3ead.jpg
│ ├── iris-5da184e8620ebf0feef4d5ffe4346e6c44b2fb60cecc0320bd7726a1844b14cd.jpg
│ ├── iris-66eee9ff0bfa521905f733b2a0c6c5acad7b8f1a30d280ed4a17f54fe1822a7e.jpg
│ ├── iris-6815050b6117cf2e1fd60b1c33bfbb94837b8e173ff869f625757da4a04965c9.jpg
│ ├── iris-793fe85ddd6a97e9c9f184ed20d1d216e48bf85aa71633eff6d27073e0825d54.jpg
│ ├── iris-850229e6293a741277eb5efaa64d03c812f007c5d0f470992a8d4cfdb902230c.jpg
│ ├── iris-86d782d20ef7a60e905e367050b0413ca566acc672bc92add0bb0304faa54cfc.jpg
│ ├── iris-875a96790adc5672e044cf9da9d2edb397627884dfe91c488ab3fb65f65c80ff.jpg
│ ├── iris-96f06136df7a415550b90e443771d0b5b0cd990b503b64cc4987f5cb6797fa9b.jpg
│ ├── iris-9a889c96a37e8927f20773783a084f31897f075353d34a304c85e53be480e72a.jpg
│ ├── iris-9e3208f4f9fedc9598ddf26f77925a1e8df9d7865a4d6e5b4f74075d558d6a5e.jpg
│ ├── iris-a7e13b6f2d7f796768d898f5f66dceefdbd566dd4406eea9f266fc16dd68a6f2.jpg
│ ├── iris-b026efb61a9e3876749536afe183d2ace078e5e29615b07ac8792ab55ba90ebc.jpg
│ ├── iris-b3c086333cb5ccb7bb66a163cf4bf449dc0f28df27d6580a35832f32fd67bfc9.jpg
│ ├── iris-b795e034b6ea08d3cd9acaa434c67aca9d17016991e8dd7d6fd19ae8f6120b77.jpg
│ ├── iris-bb4a7ad4c844987bc9dc9dfad2b363698811efe3615512997a13cd191c23febc.jpg
│ ├── iris-bd60a6ed0369df4bea1934ef52277c32757838123456a595c0f2484959553a36.jpg
│ ├── iris-c15d6019ebe17d7446ced589ef5ef7a70474d35a8b072e0edfcec850b0a106db.jpg
│ ├── iris-c45295e76c6289504921412293d5ddbe4610bb6e3b593ea9ec90958e74b73ed2.jpg
│ ├── iris-c50d481f9fa3666c2c3808806c7c2945623f9d9a6a1d93a17133c4cb1560c41c.jpg
│ ├── iris-df4206653f1ec9909434323c05bb15ded18e72587e335f8905536c34a4be3d45.jpg
│ ├── iris-e45d869cb9d443b39d59e35c2f47870f5a2a335fce53f0c8a5bc615b9c53c429.jpg
│ ├── iris-e76fa5406e02a312c102f16eb5d27c7e0de37b35f801e1ed4c28bd4caf133e7a.jpg
│ ├── iris-e8d3fd862aae1c005bcc80a73fd34b9e683634933563e7538b520f26fd315478.jpg
│ ├── iris-ea578f650069a67e5e660bb22b46c23e0a182cbfb59cdf5448cf20ce858131b6.jpg
│ ├── iris-eba0c546e9b7b3d92f0b7eb98b2914810912990789479838807993d13787a2d9.jpg
│ ├── iris-f22d4b9605e62db13072246ff6925b9cf0240461f9dfc948d154b983db4243b9.jpg
│ ├── iris-fac5f8c23d8c50658db0f4e4a074c2f7771917eb52cbdf6eda50c12889510cf4.jpg
│ └── meta.csv
└── study2_raw_images
├── iris-01cdd55ca6402713465841abddcce79a2e906e12edf95afb77c16bde4b4907dc.jpg
├── iris-02868b71ddd9b33ab795ac41609ea7b20a6e94f2543fad5d7fa11241d61feacf.jpg
├── iris-0415d2f3295db04bebc93249b685f7d7af7873faa911cd270ecd8363bd322ed5.jpg
├── iris-0c826b6f4648edf507e0cafdab53712bb6fd1f04dab453cee8db774a728dd640.jpg
├── iris-10fb9f154ead3c56ba0ab2c1ab609521c963f2326a648f82c9d7cabd178fc425.jpg
├── iris-14cbed88b0d2a929477bdf1299724f22d782e90f29ce55531f4a3d8608f7d926.jpg
├── iris-186fe29e32ee1405ddbdd36236dd7691a3c45ba78cc4c0bf11489fa09fbb1b65.jpg
├── iris-1b0b5aabd59e4c6ed1ceb54e57534d76f2f3f97e0a81800ff7ed901c35a424ab.jpg
├── iris-1d35672eb95f5b1cf14c2977eb025c246f83cdacd056115fdc93e946b56b610c.jpg
├── iris-1f941001f508ff1bd492457a90da64e52c461bfd64587a3cf7c6bf1bcb35adab.jpg
├── iris-2a09038b87009ecee5e5b4cd4cef068653809cc1e08984f193fad00f1c0df972.jpg
├── iris-308389e34b6d9a61828b339916aed7af295fdb1c7577c23fb37252937619e7e4.jpg
├── iris-30e4e56b1f170ff4863b178a0a43ea7a64fdd06c1f89a775ec4dbf5fec71e15c.jpg
├── iris-332953f4d6a355ca189e2508164b24360fc69f83304e7384ca2203ddcb7c73b5.jpg
├── iris-338fc323ed045a908fb1e8ff991255e1b8e01c967e36b054cb65edddf97b3bb0.jpg
├── iris-34a7cc16d26ba0883574e7a1c913ad50cf630e56ec08ee1113bf3584f4e40230.jpg
├── iris-360196ba36654c0d9070f95265a8a90bc224311eb34d1ab0cf851d8407d7c28e.jpg
├── iris-36132c6df6b47bda180b1daaafc7ac8a32fd7f9af83a92569da41429da49ea5b.jpg
├── iris-36f2b9282342292b67f38a55a62b0c66fa4e5bb58587f7fec90d1e93ea8c407a.jpg
├── iris-37ad07fd7b39bc377fa6e9cafdb6e0c57fb77df2c264fe631705a8436c0c2513.jpg
├── iris-3ba1625bb78e4b69b114bdafcdab64104b211d8ebadca89409e9e7ead6a0557c.jpg
├── iris-4c5d9a33327db025d9c391aeb182cbe20cfab4d4eb4ac951cc5cd15e132145d8.jpg
├── iris-522f3eb1807d015f99e66e73b19775800712890f2c7f5b777409a451fa47d532.jpg
├── iris-589fa96b9a3c2654cf08d05d3bebf4ab7bc23592d7d5a95218f9ff87612992fa.jpg
├── iris-61b71f1de04a03ce719094b65179b06e3cd80afa01622b30cda8c3e41de6bfaa.jpg
├── iris-62ef719cd70780088a4c140afae2a96c6ca9c22b72b078e3b9d25678d00b88a5.jpg
├── iris-819130af42335d4bb75bebb0d2ee2e353a89a3d518a1d2ce69842859c5668c5a.jpg
├── iris-8669e4937a2003054408afd228d99cb737e9db5088f42d292267c43a3889001a.jpg
├── iris-86c76e0f331bc62192c392cf7c3ea710d2272a8cc9928d2566a5fc4559e5dce4.jpg
├── iris-8a8bc54332a42bb35ee131d7b64e9375b4ac890632eb09e193835b838172d797.jpg
├── iris-8e9439ec7231fa3b9bc9f62a67af4e180466b32a72316600431b1ec93e63b296.jpg
├── iris-90b7d491b9a39bb5c8bb7649cce90ab7f483c2759fb55fda2d9067ac9eec7e39.jpg
├── iris-9dededf184993455c411a0ed81d6c3c55af7c610ccb55c6ae34dfac2f8bde978.jpg
├── iris-9e6ce91679c9aaceb3e9c930f11e788aacbfa8341a2a5737583c14a4d6666f3d.jpg
├── iris-a0e65269f7dc7801ac1ad8bd0c5aa547a70c7655447e921d1d4d153a9d23815e.jpg
├── iris-a445b0720254984275097c83afbdb1fe896cb010b5c662a6532ed0601ea24d7c.jpg
├── iris-a6b85bf1f3d18bbb6470440592834c2c7f081b490836392cf5f01636ee7cf658.jpg
├── iris-b005c82b844de575f0b972b9a1797b2b1fbe98c067c484a51006afc4f549ada4.jpg
├── iris-bfcf79b3b527eb64b78f9a068a1000042336e532f0f44e68f818dd13ab492a76.jpg
├── iris-c156236fb6e888764485e796f1f972bbc7ad960fe6330a7ce9182922046439c4.jpg
├── iris-d99d5fd2de5be1419cbd569570dbb6c9a6c8ec4f0a1ff5b55dc2607f6ecdca8f.jpg
├── iris-d9aae37a8fa6afdef2af170c266a597925eea935f4d070e979d565713ea62642.jpg
├── iris-dbc87fcecade2c070baaf99caf03f4f0f6e3aa977e34972383cb94d0efe8a95d.jpg
├── iris-e3d1a560d25cf573d2cbbf2fe6cd231819e998109a5cf1788d59fbb9859b3be2.jpg
├── iris-ec288bdad71388f907457db2476f12a5cb43c28cfa28d2a2077398a42b948a35.jpg
├── iris-ed5b4e072d43bc53a00a4a7f4d0f5d7c0cbd6a006e9c2d463128cedc956cb3de.jpg
├── iris-f3018a9440d17c265062d1c61475127f9952b6fe951d38fd7700402d706c0b01.jpg
├── iris-f47c5963cdbaa3238ba2d446848e8449c6af83e663f0a9216cf0baba8429b36f.jpg
├── iris-fa4b6d7e3617216104b1405cda21bf234840cd84a2c1966034caa63def2f64f0.jpg
├── iris-fc4b0cc65387ff78471659d14a78f0309a76f4c3ec641b871e40b40424255097.jpg
└── meta.csv
Our goal is to turn these directories into a validated & queryable collection that can be used alongside many other collections.
Register an artifact#
LaminDB uses the Artifact class to model files, folders & arrays in storage with their metadata. It’s a registry to manage search, queries, validation & access of storage locations.
Let’s create a Artifact record from one of the files:
artifact = ln.Artifact(
"s3://lamindb-dev-datasets/iris_studies/study0_raw_images/meta.csv"
)
artifact
💡 path in storage 's3://lamindb-dev-datasets' with key 'iris_studies/study0_raw_images/meta.csv'
Artifact(uid='o5tTv4TU4OZGSWnULdR9', key='iris_studies/study0_raw_images/meta.csv', suffix='.csv', size=4355, hash='ZpAEpN0iFYH6vjZNigic7g', hash_type='md5', visibility=1, key_is_virtual=False, storage_id=2, transform_id=1, run_id=1, created_by_id=1)
Which fields are populated when creating an artifact record?
Basic fields:
uid: universal IDkey: storage key, a relative path of the artifact instoragedescription: an optional string descriptionstorage: the storage location (the root, say, an S3 bucket or a local directory)suffix: an optional file/path suffixsize: the artifact size in byteshash: a hash useful to check for integrity and collisions (is this artifact already stored?)hash_type: the type of the hash (usually, an MD5 or SHA1 checksum)created_at: time of creationupdated_at: time of last update
Provenance-related fields:
created_by: theUserwho created the artifacttransform: theTransform(pipeline, notebook, instrument, app) that was runrun: theRunof the transform that created the artifact
For a full reference, see Artifact.
Upon .save(), artifact metadata is written to the database:
artifact.save()
Artifact(uid='o5tTv4TU4OZGSWnULdR9', key='iris_studies/study0_raw_images/meta.csv', suffix='.csv', size=4355, hash='ZpAEpN0iFYH6vjZNigic7g', hash_type='md5', visibility=1, key_is_virtual=False, updated_at=2024-05-01 18:50:25 UTC, storage_id=2, transform_id=1, run_id=1, created_by_id=1)
What happens during save?
In the database: A artifact record is inserted into the artifact registry. If the artifact record exists already, it’s updated.
In storage:
If the default storage is in the cloud,
.save()triggers an upload for a local artifact.If the artifact is already in a registered storage location, only the metadata of the record is saved to the
artifactregistry.
We can get an overview of all artifacts in the database by calling df():
ln.Artifact.df()
| uid | storage_id | key | suffix | accessor | description | version | size | hash | hash_type | n_objects | n_observations | transform_id | run_id | visibility | key_is_virtual | created_at | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||
| 1 | o5tTv4TU4OZGSWnULdR9 | 2 | iris_studies/study0_raw_images/meta.csv | .csv | None | None | None | 4355 | ZpAEpN0iFYH6vjZNigic7g | md5 | None | None | 1 | 1 | 1 | False | 2024-05-01 18:50:25.683074+00:00 | 2024-05-01 18:50:25.683102+00:00 | 1 |
View data lineage#
Because we called track(), we know that the artifact was saved in the current notebook (view_lineage()):
artifact.view_lineage()
We can also directly access its linked Transform & Run records:
artifact.transform
Transform(uid='NJvdsWWbJlZS6K79', name='Tutorial: Artifacts', key='tutorial', version='0', type='notebook', updated_at=2024-05-01 18:50:22 UTC, created_by_id=1)
artifact.run
Run(uid='gYXIXnIw7o7ys3cRQgEl', started_at=2024-05-01 18:50:22 UTC, is_consecutive=True, transform_id=1, created_by_id=1)
(For a comprehensive example with data lineage through app uploads, pipelines & notebooks of multiple data types, see Project flow.)
Access an artifact#
path gives you the file path (UPath):
artifact.path
S3Path('s3://lamindb-dev-datasets/iris_studies/study0_raw_images/meta.csv')
To cache the artifact to a local cache, call cache():
artifact.cache()
PosixUPath('/home/runner/.cache/lamindb/lamindb-dev-datasets/iris_studies/study0_raw_images/meta.csv')
To load data into memory with a default loader, call load():
df = artifact.load(index_col=0)
df.head()
| 0 | 1 | |
|---|---|---|
| 0 | iris-0797945218a97d6e5251b4758a2ba1b418cbd52ce... | setosa |
| 1 | iris-0f133861ea3fe1b68f9f1b59ebd9116ff963ee710... | versicolor |
| 2 | iris-9ffe51c2abd973d25a299647fa9ccaf6aa9c8eecf... | versicolor |
| 3 | iris-83f433381b755101b9fc9fbc9743e35fbb8a1a109... | setosa |
| 4 | iris-bdae8314e4385d8e2322abd8e63a82758a9063c77... | virginica |
If the data is large, you’ll likely want to query it via backed(). For more on this, see: Query arrays.
How do I update an artifact?
If you’d like to replace the underlying stored object, use replace().
If you’d like to update metadata:
artifact.description = "My new description"
artifact.save() # save the change to the database
Register directories as artifacts#
We now register the entire directory for study 0 as an artifact:
study0_data = ln.Artifact(f"s3://lamindb-dev-datasets/iris_studies/study0_raw_images")
study0_data.save()
ln.Artifact.df() # see the registry content
💡 path in storage 's3://lamindb-dev-datasets' with key 'iris_studies/study0_raw_images'
| uid | storage_id | key | suffix | accessor | description | version | size | hash | hash_type | n_objects | n_observations | transform_id | run_id | visibility | key_is_virtual | created_at | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||
| 2 | 7zPAagBwZTJzQqVLFyqB | 2 | iris_studies/study0_raw_images | None | None | None | 656692 | wVYKPpEsmmrqSpAZIRXCFg | md5-d | 51.0 | None | 1 | 1 | 1 | False | 2024-05-01 18:50:26.600312+00:00 | 2024-05-01 18:50:26.600341+00:00 | 1 | |
| 1 | o5tTv4TU4OZGSWnULdR9 | 2 | iris_studies/study0_raw_images/meta.csv | .csv | None | None | None | 4355 | ZpAEpN0iFYH6vjZNigic7g | md5 | NaN | None | 1 | 1 | 1 | False | 2024-05-01 18:50:25.683074+00:00 | 2024-05-01 18:50:25.683102+00:00 | 1 |
Filter & search artifacts#
You can search artifacts directly based on the Artifact registry:
ln.Artifact.search("meta").head()
| key | description | score | |
|---|---|---|---|
| uid | |||
| o5tTv4TU4OZGSWnULdR9 | iris_studies/study0_raw_images/meta.csv | 60.0 | |
| 7zPAagBwZTJzQqVLFyqB | iris_studies/study0_raw_images | 48.9 |
You can also query & search the artifact by any metadata combination.
For instance, look up a user with auto-complete from the User registry:
users = ln.User.lookup()
users.anonymous
User(uid='00000000', handle='anonymous', updated_at=2024-05-01 18:50:20 UTC)
How do I act non-anonymously?
Filter the Transform registry for a name:
transform = ln.Transform.filter(
name__icontains="Artifacts"
).one() # get exactly one result
transform
Transform(uid='NJvdsWWbJlZS6K79', name='Tutorial: Artifacts', key='tutorial', version='0', type='notebook', updated_at=2024-05-01 18:50:22 UTC, created_by_id=1)
What does a double underscore mean?
For any field, the double underscore defines a comparator, e.g.,
name__icontains="Martha":namecontains"Martha"when ignoring casename__startswith="Martha":namestarts with"Marthaname__in=["Martha", "John"]:nameis"John"or"Martha"
For more info, see: Query & search registries.
Use these results to filter the Artifact registry:
ln.Artifact.filter(
created_by=users.anonymous,
transform=transform,
suffix=".csv",
).df().head()
| uid | storage_id | key | suffix | accessor | description | version | size | hash | hash_type | n_objects | n_observations | transform_id | run_id | visibility | key_is_virtual | created_at | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||
| 1 | o5tTv4TU4OZGSWnULdR9 | 2 | iris_studies/study0_raw_images/meta.csv | .csv | None | None | None | 4355 | ZpAEpN0iFYH6vjZNigic7g | md5 | None | None | 1 | 1 | 1 | False | 2024-05-01 18:50:25.683074+00:00 | 2024-05-01 18:50:25.683102+00:00 | 1 |
You can also query for directories using key__startswith (LaminDB treats directories like AWS S3, as the prefix of the storage key):
ln.Artifact.filter(key__startswith="iris_studies/study0_raw_images/").df().head()
| uid | storage_id | key | suffix | accessor | description | version | size | hash | hash_type | n_objects | n_observations | transform_id | run_id | visibility | key_is_virtual | created_at | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||
| 1 | o5tTv4TU4OZGSWnULdR9 | 2 | iris_studies/study0_raw_images/meta.csv | .csv | None | None | None | 4355 | ZpAEpN0iFYH6vjZNigic7g | md5 | None | None | 1 | 1 | 1 | False | 2024-05-01 18:50:25.683074+00:00 | 2024-05-01 18:50:25.683102+00:00 | 1 |
Note
You can look up, filter & search any registry (Registry).
You can chain filter() statements and search(): ln.Artifact.filter(suffix=".jpg").search("my image")
An empty filter returns the entire registry: ln.Artifact.filter()
For more info, see: Query & search registries.
Filter & search on LaminHub
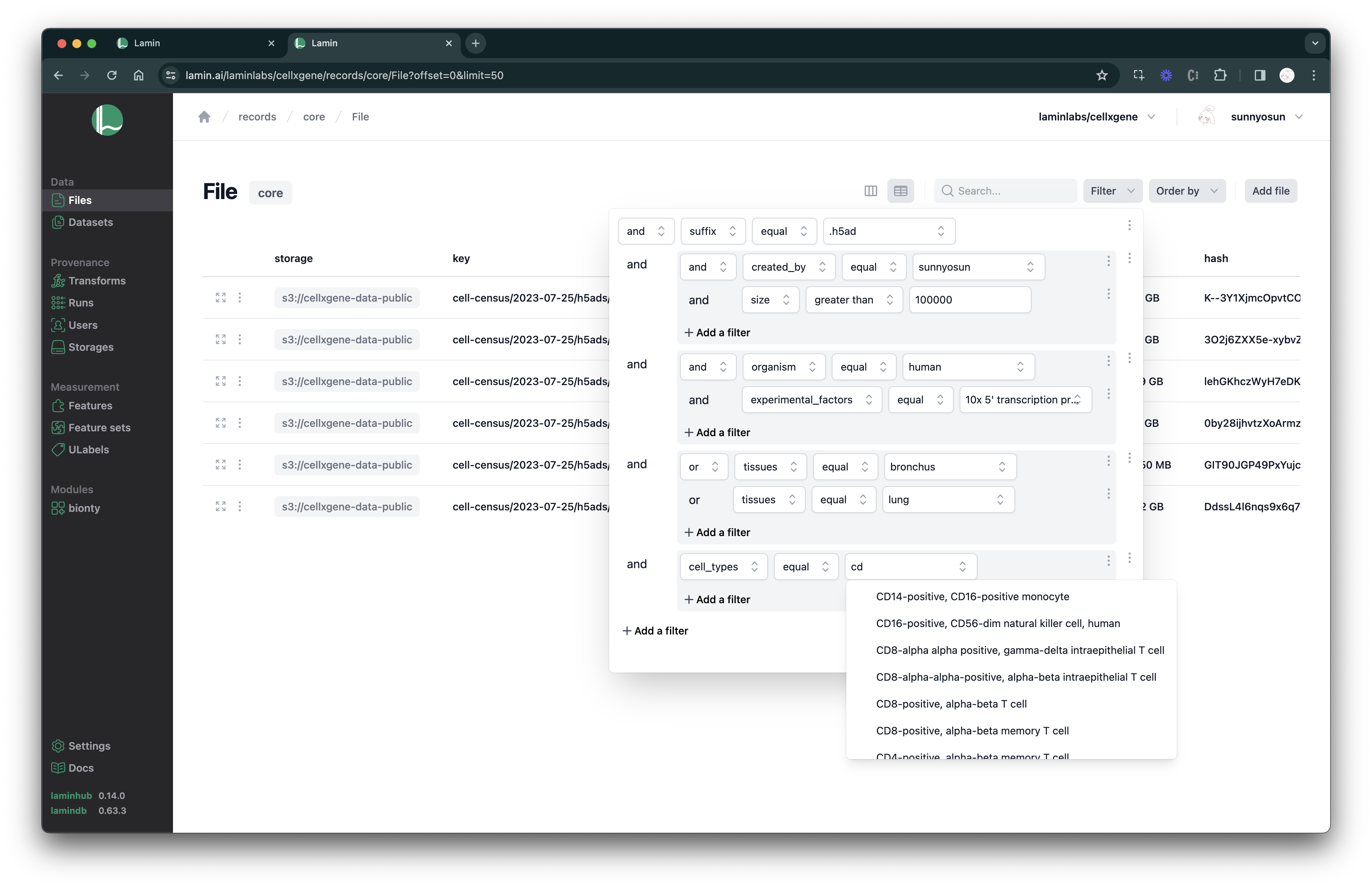
Describe artifacts#
Get an overview of what happened:
artifact.describe()
Artifact(uid='o5tTv4TU4OZGSWnULdR9', key='iris_studies/study0_raw_images/meta.csv', suffix='.csv', size=4355, hash='ZpAEpN0iFYH6vjZNigic7g', hash_type='md5', visibility=1, key_is_virtual=False, updated_at=2024-05-01 18:50:25 UTC)
Provenance:
📎 storage: Storage(uid='EzvEnPnH', root='s3://lamindb-dev-datasets', type='s3', region='us-east-1', instance_uid='pZ1VQkyD3haH')
📎 transform: Transform(uid='NJvdsWWbJlZS6K79', name='Tutorial: Artifacts', key='tutorial', version='0', type='notebook')
📎 run: Run(uid='gYXIXnIw7o7ys3cRQgEl', started_at=2024-05-01 18:50:22 UTC, is_consecutive=True)
📎 created_by: User(uid='00000000', handle='anonymous')
artifact.view_lineage()
Version artifacts#
If you’d like to version an artifact or transform, either provide the version parameter when creating it or create new versions through is_new_version_of.
For instance:
new_artifact = ln.Artifact(data, is_new_version_of=old_artifact)
If you’d like to add a registered artifact to a version family, use add_to_version_family.
For instance:
new_artifact.add_to_version_family(old_artifact)
Are there remaining questions about storing artifacts? If so, see: Storage FAQ.
Collections#
An artifact can model anything that’s in storage: a file, a collection, an array, a machine learning model.
Often times, several artifacts together represent a collection.
Let’s store the artifact for study0_data as a Collection:
collection = ln.Collection(
study0_data,
name="Iris collection",
version="1",
description="50 image files and metadata",
)
collection
Collection(uid='32cBtnkazA7BtJB4YV3x', name='Iris collection', description='50 image files and metadata', version='1', hash='WwFLpNFmK8GMC2dSGj1W', visibility=1, transform_id=1, run_id=1, created_by_id=1)
And save it:
collection.save()
Now, we perform subsequent studies by collecting more data.
We’d like to keep track of their data as part of a growing versioned collection:
artifacts = [study0_data]
for folder_name in ["study1_raw_images", "study2_raw_images"]:
# create an artifact for the folder
artifact = ln.Artifact(f"s3://lamindb-dev-datasets/iris_studies/{folder_name}")
artifact.save()
artifacts.append(artifact)
# create a new version of the collection
collection = ln.Collection(
artifacts, is_new_version_of=collection, description="Another 50 images"
)
collection.description = "Another 50 images"
collection.save()
💡 path in storage 's3://lamindb-dev-datasets' with key 'iris_studies/study1_raw_images'
💡 path in storage 's3://lamindb-dev-datasets' with key 'iris_studies/study2_raw_images'
See all artifacts:
ln.Artifact.df()
| uid | storage_id | key | suffix | accessor | description | version | size | hash | hash_type | n_objects | n_observations | transform_id | run_id | visibility | key_is_virtual | created_at | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||
| 4 | KN5tE6KT9QdtP29X8w5C | 2 | iris_studies/study2_raw_images | None | None | None | 665518 | PX8Vt9T28y-uCEJO1tKm7A | md5-d | 51.0 | None | 1 | 1 | 1 | False | 2024-05-01 18:50:28.003536+00:00 | 2024-05-01 18:50:28.003564+00:00 | 1 | |
| 3 | 3ksHD7VWCbUJld5fiYzd | 2 | iris_studies/study1_raw_images | None | None | None | 640617 | j61W__GgImA18CKrIf7FVg | md5-d | 49.0 | None | 1 | 1 | 1 | False | 2024-05-01 18:50:27.385608+00:00 | 2024-05-01 18:50:27.385637+00:00 | 1 | |
| 2 | 7zPAagBwZTJzQqVLFyqB | 2 | iris_studies/study0_raw_images | None | None | None | 656692 | wVYKPpEsmmrqSpAZIRXCFg | md5-d | 51.0 | None | 1 | 1 | 1 | False | 2024-05-01 18:50:26.600312+00:00 | 2024-05-01 18:50:26.600341+00:00 | 1 | |
| 1 | o5tTv4TU4OZGSWnULdR9 | 2 | iris_studies/study0_raw_images/meta.csv | .csv | None | None | None | 4355 | ZpAEpN0iFYH6vjZNigic7g | md5 | NaN | None | 1 | 1 | 1 | False | 2024-05-01 18:50:25.683074+00:00 | 2024-05-01 18:50:25.683102+00:00 | 1 |
See all collections:
ln.Collection.df()
| uid | name | description | version | hash | reference | reference_type | transform_id | run_id | artifact_id | visibility | created_at | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||
| 3 | 32cBtnkazA7BtJB4pisU | Iris collection | Another 50 images | 3 | T-U8z2Zi5rFYdAD9pzmS | None | None | 1 | 1 | None | 1 | 2024-05-01 18:50:28.016434+00:00 | 2024-05-01 18:50:28.016457+00:00 | 1 |
| 2 | 32cBtnkazA7BtJB4Db1u | Iris collection | Another 50 images | 2 | 5cCK6ZLOPB0cV3tyeZup | None | None | 1 | 1 | None | 1 | 2024-05-01 18:50:27.398726+00:00 | 2024-05-01 18:50:27.398750+00:00 | 1 |
| 1 | 32cBtnkazA7BtJB4YV3x | Iris collection | 50 image files and metadata | 1 | WwFLpNFmK8GMC2dSGj1W | None | None | 1 | 1 | None | 1 | 2024-05-01 18:50:26.800334+00:00 | 2024-05-01 18:50:26.800363+00:00 | 1 |
Most functionality that you just learned about artifacts - e.g., queries & provenance - also applies to Collection.
But Collection is an abstraction over storing data in one or several artifacts and does not have a key field.
We’ll learn more about collections in the next part of the tutorial.
View changes#
With view(), you can see the latest changes to the database:
ln.view() # link tables in the database are not shown
Show code cell output
Artifact
| uid | storage_id | key | suffix | accessor | description | version | size | hash | hash_type | n_objects | n_observations | transform_id | run_id | visibility | key_is_virtual | created_at | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||
| 4 | KN5tE6KT9QdtP29X8w5C | 2 | iris_studies/study2_raw_images | None | None | None | 665518 | PX8Vt9T28y-uCEJO1tKm7A | md5-d | 51.0 | None | 1 | 1 | 1 | False | 2024-05-01 18:50:28.003536+00:00 | 2024-05-01 18:50:28.003564+00:00 | 1 | |
| 3 | 3ksHD7VWCbUJld5fiYzd | 2 | iris_studies/study1_raw_images | None | None | None | 640617 | j61W__GgImA18CKrIf7FVg | md5-d | 49.0 | None | 1 | 1 | 1 | False | 2024-05-01 18:50:27.385608+00:00 | 2024-05-01 18:50:27.385637+00:00 | 1 | |
| 2 | 7zPAagBwZTJzQqVLFyqB | 2 | iris_studies/study0_raw_images | None | None | None | 656692 | wVYKPpEsmmrqSpAZIRXCFg | md5-d | 51.0 | None | 1 | 1 | 1 | False | 2024-05-01 18:50:26.600312+00:00 | 2024-05-01 18:50:26.600341+00:00 | 1 | |
| 1 | o5tTv4TU4OZGSWnULdR9 | 2 | iris_studies/study0_raw_images/meta.csv | .csv | None | None | None | 4355 | ZpAEpN0iFYH6vjZNigic7g | md5 | NaN | None | 1 | 1 | 1 | False | 2024-05-01 18:50:25.683074+00:00 | 2024-05-01 18:50:25.683102+00:00 | 1 |
Collection
| uid | name | description | version | hash | reference | reference_type | transform_id | run_id | artifact_id | visibility | created_at | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||
| 3 | 32cBtnkazA7BtJB4pisU | Iris collection | Another 50 images | 3 | T-U8z2Zi5rFYdAD9pzmS | None | None | 1 | 1 | None | 1 | 2024-05-01 18:50:28.016434+00:00 | 2024-05-01 18:50:28.016457+00:00 | 1 |
| 2 | 32cBtnkazA7BtJB4Db1u | Iris collection | Another 50 images | 2 | 5cCK6ZLOPB0cV3tyeZup | None | None | 1 | 1 | None | 1 | 2024-05-01 18:50:27.398726+00:00 | 2024-05-01 18:50:27.398750+00:00 | 1 |
| 1 | 32cBtnkazA7BtJB4YV3x | Iris collection | 50 image files and metadata | 1 | WwFLpNFmK8GMC2dSGj1W | None | None | 1 | 1 | None | 1 | 2024-05-01 18:50:26.800334+00:00 | 2024-05-01 18:50:26.800363+00:00 | 1 |
Run
| uid | transform_id | started_at | finished_at | created_by_id | json | report_id | environment_id | is_consecutive | reference | reference_type | created_at | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||
| 1 | gYXIXnIw7o7ys3cRQgEl | 1 | 2024-05-01 18:50:22.948429+00:00 | None | 1 | None | None | None | True | None | None | 2024-05-01 18:50:22.948563+00:00 |
Storage
| uid | root | description | type | region | instance_uid | created_at | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||
| 2 | EzvEnPnH | s3://lamindb-dev-datasets | None | s3 | us-east-1 | pZ1VQkyD3haH | 2024-05-01 18:50:25.627040+00:00 | 2024-05-01 18:50:25.627073+00:00 | 1 |
| 1 | sXnIoPYEDMeP | /home/runner/work/lamindb/lamindb/docs/lamin-t... | None | local | None | 5WuFt3cW4zRx | 2024-05-01 18:50:20.106918+00:00 | 2024-05-01 18:50:20.106942+00:00 | 1 |
Transform
| uid | name | key | version | description | type | latest_report_id | source_code_id | reference | reference_type | created_at | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||
| 1 | NJvdsWWbJlZS6K79 | Tutorial: Artifacts | tutorial | 0 | None | notebook | None | None | None | None | 2024-05-01 18:50:22.941759+00:00 | 2024-05-01 18:50:22.941802+00:00 | 1 |
User
| uid | handle | name | created_at | updated_at | |
|---|---|---|---|---|---|
| id | |||||
| 1 | 00000000 | anonymous | None | 2024-05-01 18:50:20.102450+00:00 | 2024-05-01 18:50:20.102474+00:00 |
Save notebook & scripts#
When you’ve completed the work on a notebook or script, you can save the source code and, for notebooks, an execution report to your storage location like so:
ln.finish()
This enables you to query execution report & source code via transform.latest_report and transform.source_code.
If you registered the instance on LaminHub, you can share it like here.
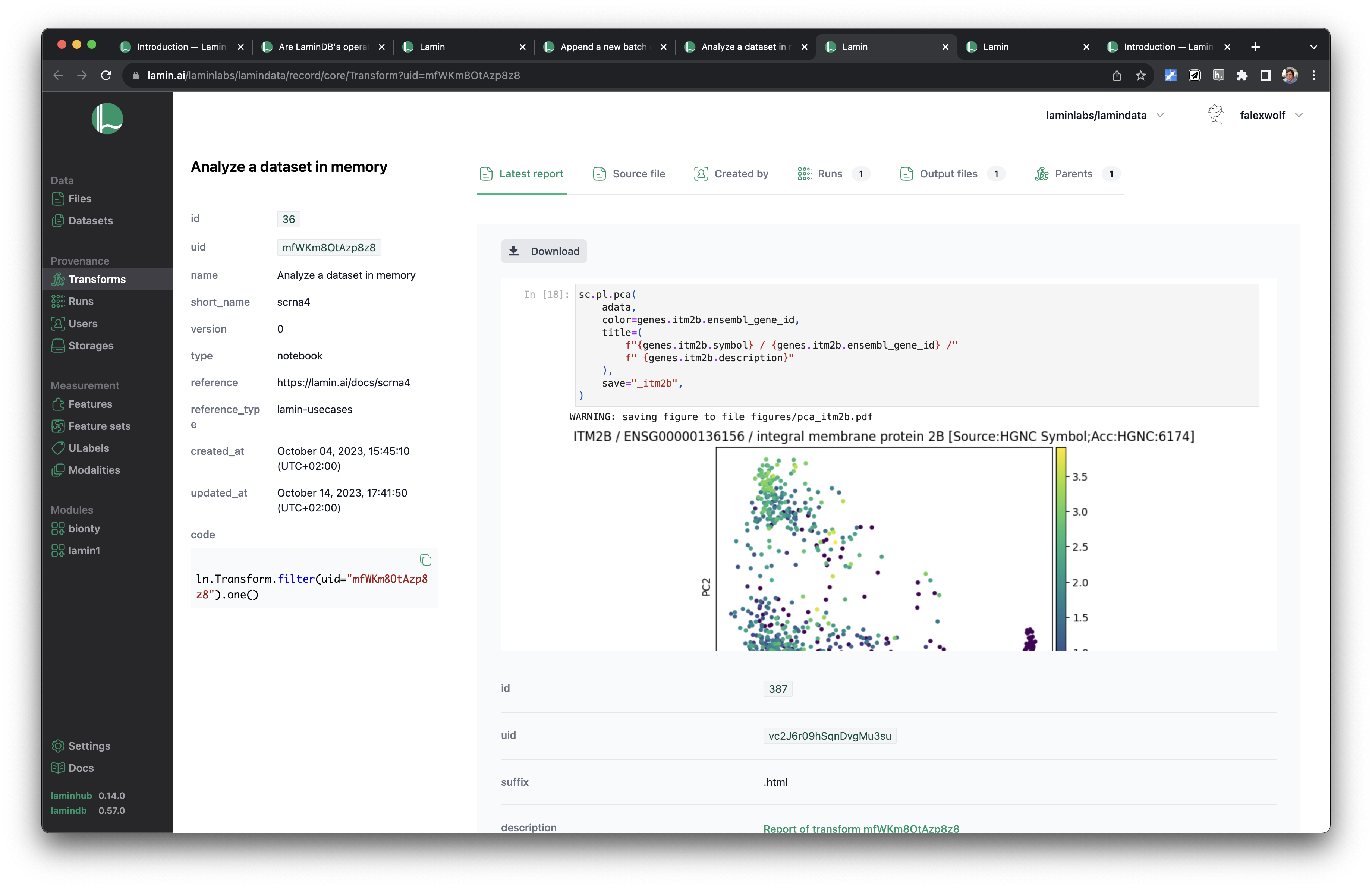
If you want to cache a notebook or script, call:
lamin get https://lamin.ai/laminlabs/lamindata/transform/NJvdsWWbJlZSz8
Read on#
Now, you already know about 6 out of 9 LaminDB core classes! The two most central are:
And the four registries related to provenance:
Transform: transforms of artifactsRun: runs of transformsUser: usersStorage: storage locations like S3/GCP buckets or local directories
If you want to validate data, label artifacts, and manage features, read on: Tutorial: Features & labels.


