Features#
LaminDB#
Access data & metadata across storage (files, arrays) & database (SQL) backends.
Manage
Feature,FeatureSet,ULabelPlug-in custom schemas & manage schema migrations
Use array formats in memory & storage: DataFrame, AnnData, MuData, SOMA, … backed by parquet, zarr, TileDB, HDF5, h5ad, DuckDB, …
Bridge artifacts and warehousing:
Artifact,CollectionLeverage out-of-the-box PyTorch data loaders:
mapped()Version artifacts, collections & transforms
Track data lineage across notebooks, pipelines & UI: track(), Transform & Run.
Execution reports, source code and Python environments for notebooks & scripts
Integrate with workflow managers: redun, nextflow, snakemake
Manage registries for experimental metadata & in-house ontologies, import public ontologies.
Use >20 public ontologies with plug-in
biontySafeguards against typos & duplications
Validate, standardize & annotate based on registries: validate & standardize.
Inspect validation failures:
inspectAnnotate with untyped or typed labels:
addSave data & metadata ACID:
save
Organize and share data across a mesh of LaminDB instances.
Create & load instances like git repos:
lamin init&lamin loadZero-copy transfer data across instances
Zero lock-in, scalable, auditable, access management, and more.
Zero lock-in: LaminDB runs on generic backends server-side and is not a client for “Lamin Cloud”
Flexible storage backends (local, S3, GCP, anything fsspec supports)
Currently two SQL backends for managing metadata: SQLite & Postgres
Scalable: metadata tables support 100s of millions of entries
Auditable: data & metadata records are hashed, timestamped, and attributed to users (soon to come: LaminDB Log)
Access management:
High-level access management through Lamin’s collaborator roles
Fine-grained access management via storage & SQL roles (soon to come: Lamin Vault)
Secure: embedded in your infrastructure (Lamin has no access to your data & metadata)
Tested & typed (up to Django Model fields)
Idempotent & ACID operations
LaminHub#
Secure & intuitive access management.
LaminHub provides a layer for AWS & GCP that makes access management more secure & intuitive.
Rather than configuring storage & database permissions directly on AWS or GCP, LaminHub allows you to manage collaborators for databases & storages as intuitively as you manage access to git repositories on GitHub.
In contrast to a closed SaaS product like GitHub, LaminHub leaves you in full control of your data with direct API access to storage & databases on AWS or GCP.
How does it work? Based on an identity provider (Google, GitHub, SSO, OIDC) and a role-based permission system, LaminDB users automatically receive federated access tokens for resources on AWS or GCP.
A UI to work with LaminDB instances.
Explore in the hub UI or lamin load owner/instance via the CLI:
lamin.ai/laminlabs/arrayloader-benchmarks - Work with ML models & benchmarks
lamin.ai/laminlabs/cellxgene - An instance with the CELLxGENE data (guide)
lamin.ai/laminlabs/lamindata - A generic demo instance with various data types
See validated datasets in context of ontologies & experimental metadata.
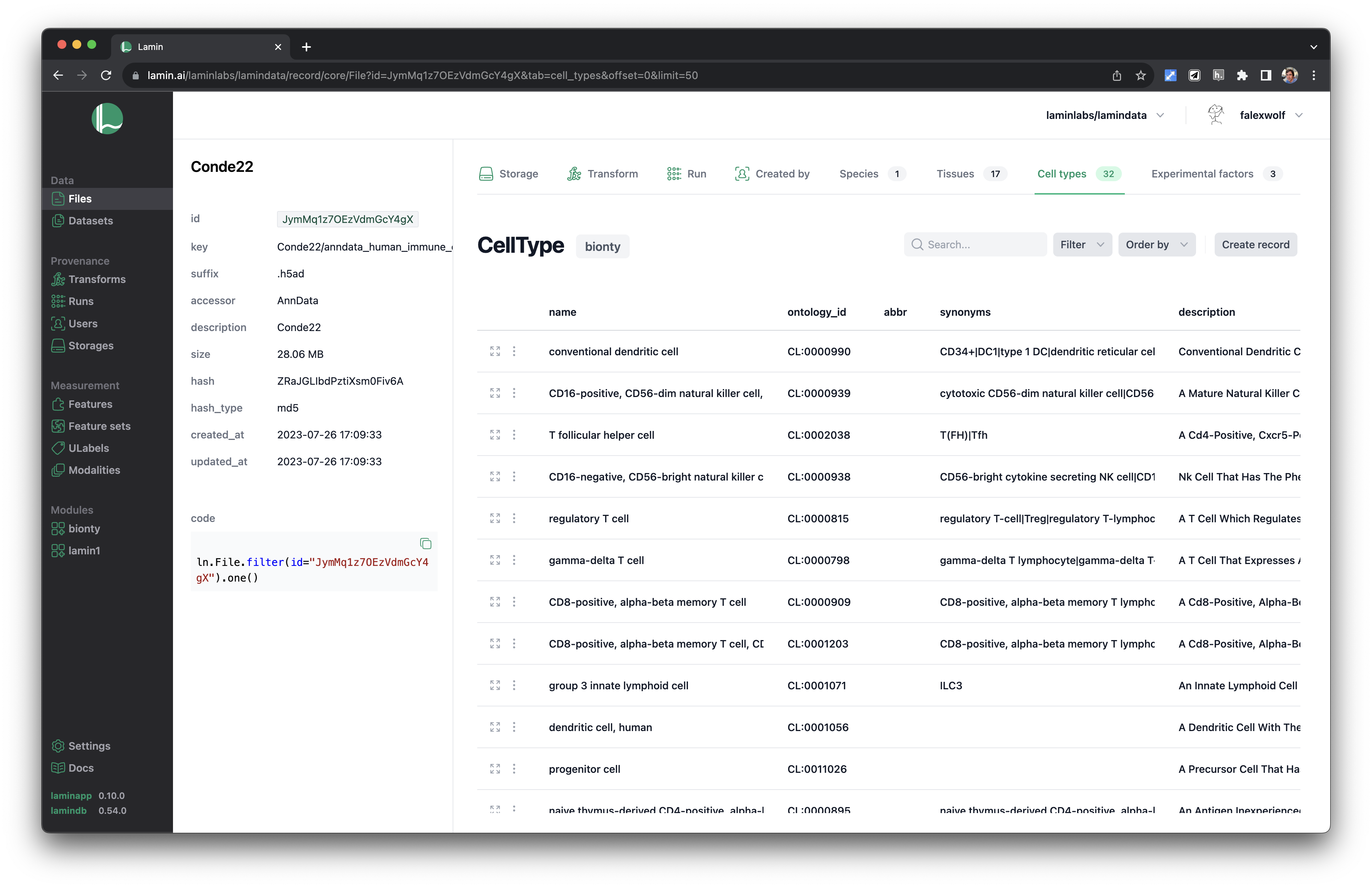
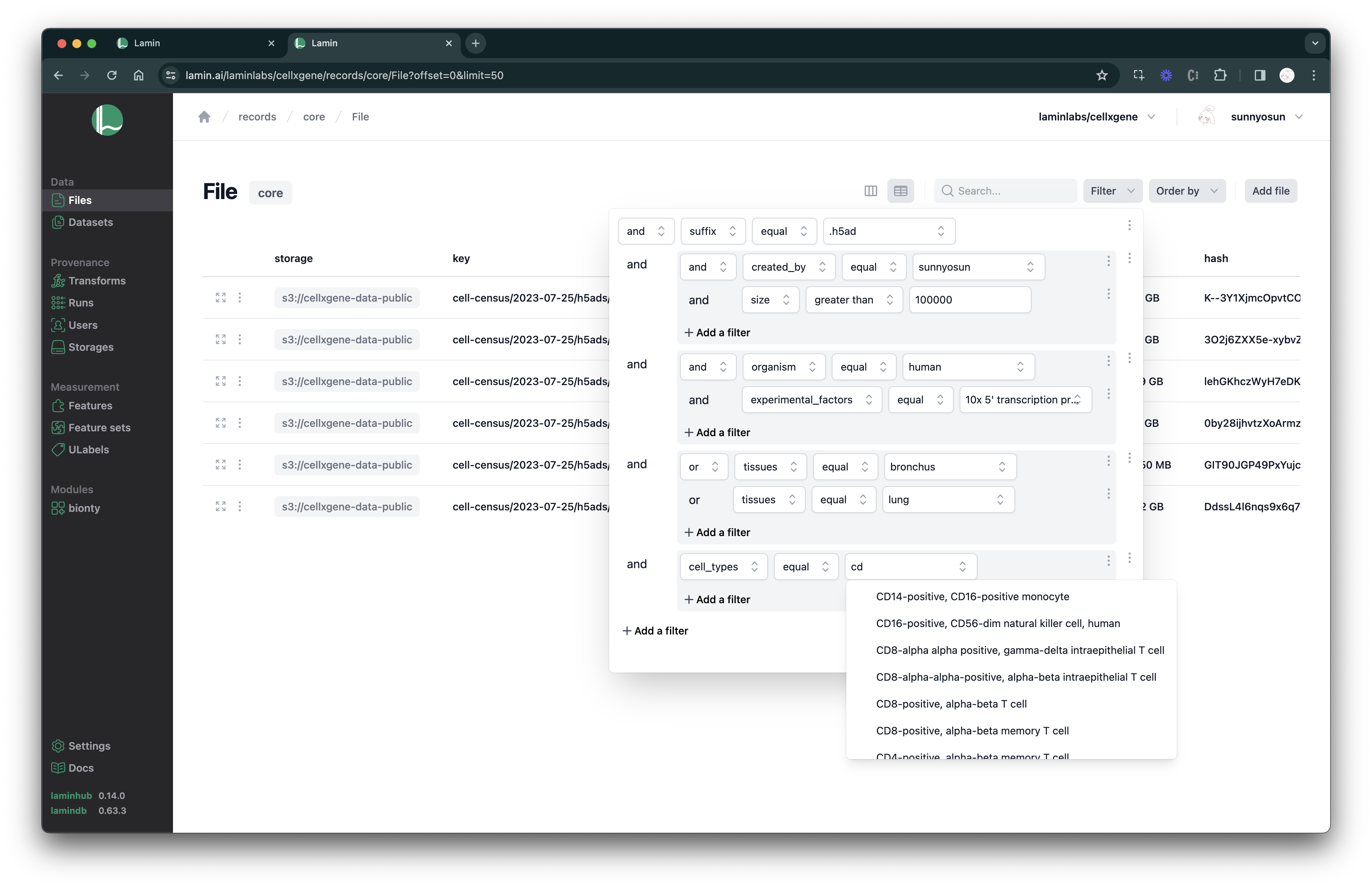
Query & search.
See scripts, notebooks & pipelines with their inputs & outputs.
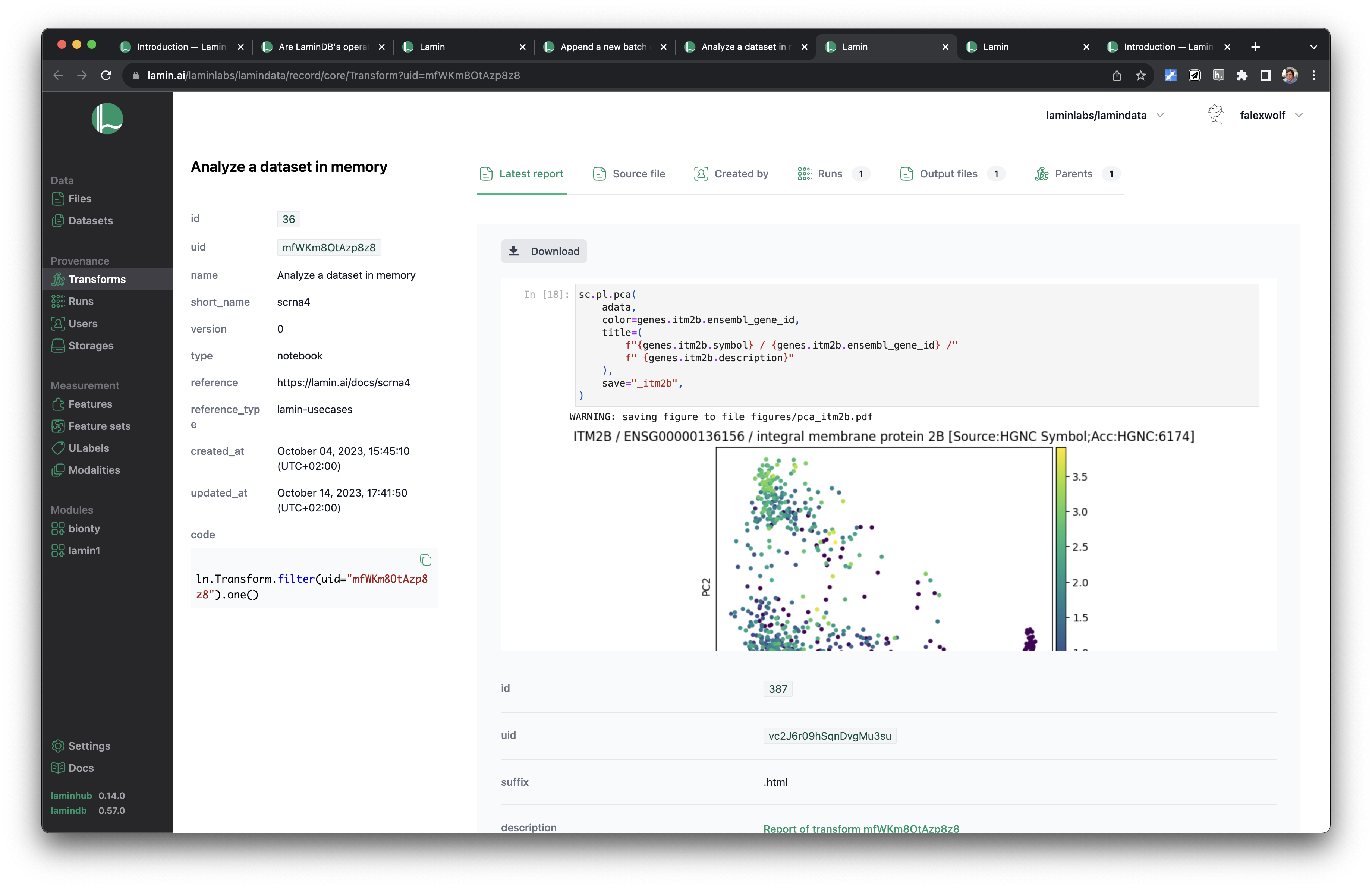
Track pipelines, notebooks & UI transforms in one registry.
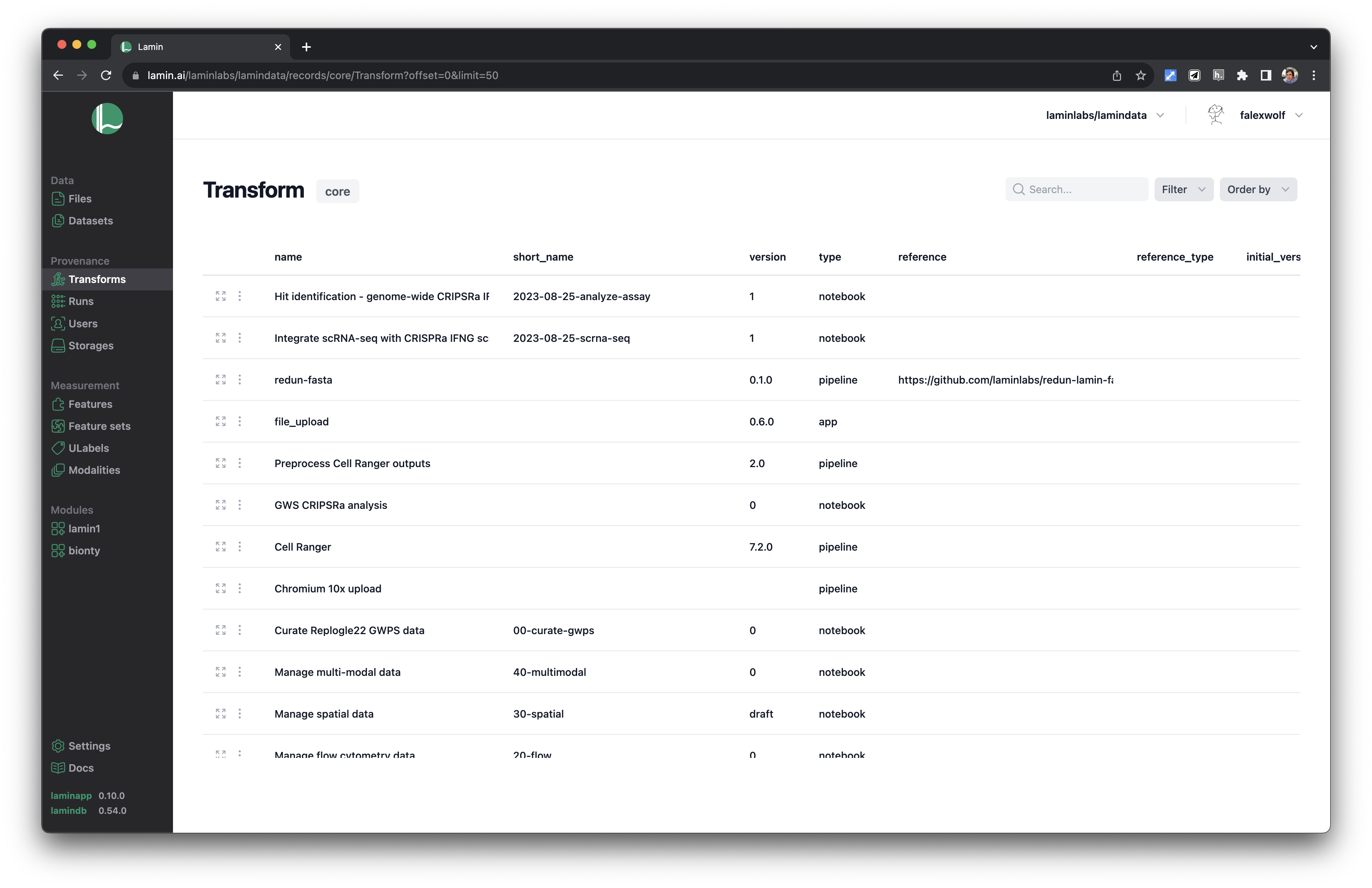
See parents and children of transforms.